4UTQ
 
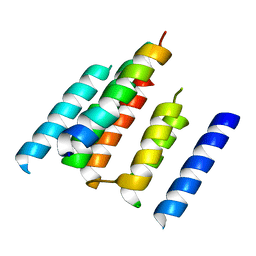 | | A structural model of the active ribosome-bound membrane protein insertase YidC | | Descriptor: | ATP SYNTHASE SUBUNIT C, MEMBRANE PROTEIN INSERTASE YIDC | | Authors: | Wickles, S, Singharoy, A, Andreani, J, Seemayer, S, Bischoff, L, Berninghausen, O, Soeding, J, Schulten, K, vanderSluis, E.O, Beckmann, R. | | Deposit date: | 2014-07-22 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | A Structural Model of the Active Ribosome-Bound Membrane Protein Insertase Yidc.
Elife, 3, 2014
|
|
7ZWA
 
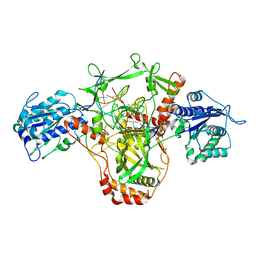 | | CryoEM structure of Ku heterodimer bound to DNA and PAXX | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*T)-3'), PHOSPHATE ION, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
7ZYG
 
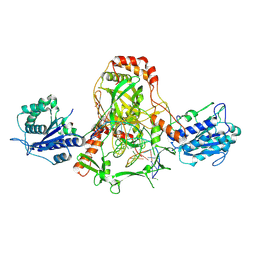 | | CryoEM structure of Ku heterodimer bound to DNA, PAXX and XLF | | Descriptor: | DNA, Non-homologous end-joining factor 1, PHOSPHATE ION, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
7ZVT
 
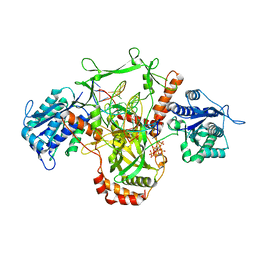 | | CryoEM structure of Ku heterodimer bound to DNA | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*TP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-17 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
4AM3
 
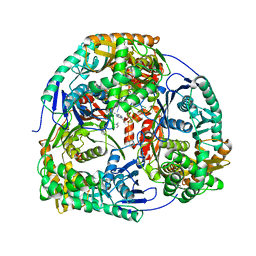 | | Crystal structure of C. crescentus PNPase bound to RNA | | Descriptor: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RNA, ... | | Authors: | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | Deposit date: | 2012-03-07 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
4AIM
 
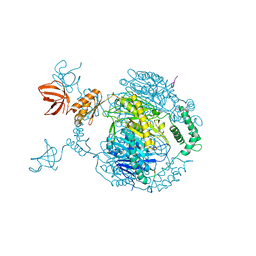 | | Crystal structure of C. crescentus PNPase bound to RNase E recognition peptide | | Descriptor: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RIBONUCLEASE, ... | | Authors: | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | Deposit date: | 2012-02-10 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
4AID
 
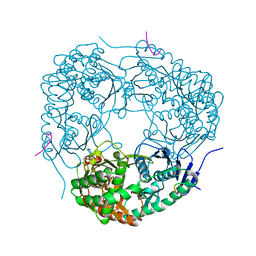 | | Crystal structure of C. crescentus PNPase bound to RNase E recognition peptide | | Descriptor: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RIBONUCLEASE, ... | | Authors: | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | Deposit date: | 2012-02-09 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
1BJ4
 
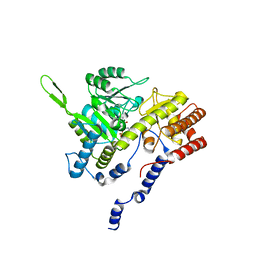 | | RECOMBINANT SERINE HYDROXYMETHYLTRANSFERASE (HUMAN) | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, cytosolic | | Authors: | Renwick, S.B, Snell, K. | | Deposit date: | 1998-07-02 | | Release date: | 1999-08-16 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The crystal structure of human cytosolic serine hydroxymethyltransferase: a target for cancer chemotherapy
Structure, 6, 1998
|
|
8BOT
 
 | |
8BH3
 
 | | DNA-PK Ku80 mediated dimer bound to PAXX | | Descriptor: | DNA (25-MER), DNA (26-MER), DNA (27-MER), ... | | Authors: | Hardwick, S.W, Chaplin, A.K. | | Deposit date: | 2022-10-28 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
8BHY
 
 | |
8BHV
 
 | | DNA-PK XLF mediated dimer bound to PAXX | | Descriptor: | DNA (24-MER), DNA (26-MER), DNA (27-MER), ... | | Authors: | Hardwick, S.W, Chaplin, A.K. | | Deposit date: | 2022-11-01 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.51 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
1BLX
 
 | | P19INK4D/CDK6 COMPLEX | | Descriptor: | CALCIUM ION, CYCLIN-DEPENDENT KINASE 6, P19INK4D | | Authors: | Brotherton, D.H, Dhanaraj, V, Wick, S, Brizuela, L, Domaille, P.J, Volyanik, E, Xu, X, Parisini, E, Smith, B.O, Archer, S.J, Serrano, M, Brenner, S.L, Blundell, T.L, Laue, E.D. | | Deposit date: | 1998-07-21 | | Release date: | 1999-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of the cyclin D-dependent kinase Cdk6 bound to the cell-cycle inhibitor p19INK4d.
Nature, 395, 1998
|
|
5FT0
 
 | | Crystal structure of gp37(Dip) from bacteriophage phiKZ | | Descriptor: | ARGININE, GP37, POTASSIUM ION | | Authors: | Van den Bossche, A, Hardwick, S.W, Ceyssens, P.J, Hendrix, H, Voet, M, Dendooven, T, Bandyra, K.J, De Maeyer, M, Aertsen, A, Noben, J.P, Luisi, B.F, Lavigne, R. | | Deposit date: | 2016-01-08 | | Release date: | 2016-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural elucidation of a novel mechanism for the bacteriophage-based inhibition of the RNA degradosome.
Elife, 5, 2016
|
|
5FT1
 
 | | Crystal structure of gp37(Dip) from bacteriophage phiKZ bound to RNase E of Pseudomonas aeruginosa | | Descriptor: | GP37, RIBONUCLEASE E | | Authors: | Van den Bossche, A, Hardwick, S.W, Ceyssens, P.J, Hendrix, H, Voet, M, Dendooven, T, Bandyra, K.J, De Maeyer, M, Aertsen, A, Noben, J.P, Luisi, B.F, Lavigne, R. | | Deposit date: | 2016-01-08 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural elucidation of a novel mechanism for the bacteriophage-based inhibition of the RNA degradosome.
Elife, 5, 2016
|
|
4GG2
 
 | | The crystal structure of glutamate-bound human gamma-glutamyltranspeptidase 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | West, M.B, Chen, Y, Wickham, S, Heroux, A, Cahill, K, Hanigan, M.H, Mooers, B.H.M. | | Deposit date: | 2012-08-04 | | Release date: | 2013-09-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Novel Insights into Eukaryotic gamma-Glutamyltranspeptidase 1 from the Crystal Structure of the Glutamate-bound Human Enzyme.
J.Biol.Chem., 288, 2013
|
|
3J15
 
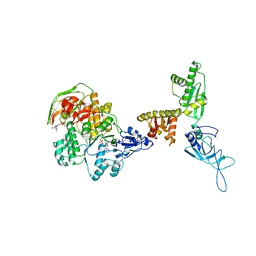 | | Model of ribosome-bound archaeal Pelota and ABCE1 | | Descriptor: | ABC transporter ATP-binding protein, ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, ... | | Authors: | Becker, T, Franckenberg, S, Wickles, S, Shoemaker, C.J, Anger, A.M, Armache, J.-P, Sieber, H, Ungewickell, C, Berninghausen, O, Daberkow, I, Karcher, A, Thomm, M, Hopfner, K.-P, Green, R, Beckmann, R. | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural basis of highly conserved ribosome recycling in eukaryotes and archaea.
Nature, 482, 2012
|
|
3J16
 
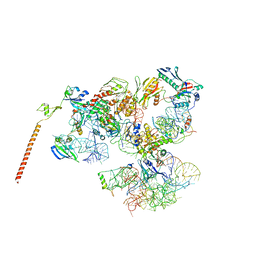 | | Models of ribosome-bound Dom34p and Rli1p and their ribosomal binding partners | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S24-A, ... | | Authors: | Becker, T, Franckenberg, S, Wickles, S, Shoemaker, C.J, Anger, A.M, Armache, J.-P, Sieber, H, Ungewickell, C, Berninghausen, O, Daberkow, I, Karcher, A, Thomm, M, Hopfner, K.-P, Green, R, Beckmann, R. | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structural basis of highly conserved ribosome recycling in eukaryotes and archaea.
Nature, 482, 2012
|
|
7NZM
 
 | | Cryo-EM structure of pre-dephosphorylation complex of phosphorylated eIF2alpha with trapped holophosphatase (PP1A_D64A/PPP1R15A/G-actin/DNase I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yan, Y, Hardwick, S, Ron, D. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Higher-order phosphatase-substrate contacts terminate the integrated stress response.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4OXP
 
 | |
5M5H
 
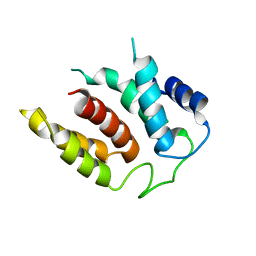 | | RIBOSOME-BOUND YIDC INSERTASE | | Descriptor: | Membrane protein insertase YidC | | Authors: | Kedrov, A, Wickles, S, Crevenna, A.H, van der Sluis, E, Buschauer, R, Berninghausen, O, Lamb, D.C, Beckmann, R. | | Deposit date: | 2016-10-21 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural Dynamics of the YidC:Ribosome Complex during Membrane Protein Biogenesis.
Cell Rep, 17, 2016
|
|
1MUO
 
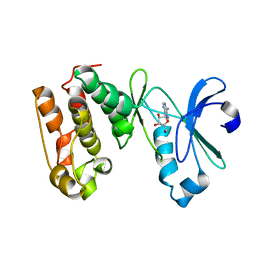 | | CRYSTAL STRUCTURE OF AURORA-2, AN ONCOGENIC SERINE-THREONINE KINASE | | Descriptor: | ADENOSINE, Aurora-related kinase 1 | | Authors: | Cheetham, G.M.T, Knegtel, R.M.A, Coll, J.T, Renwick, S.B, Swenson, L, Weber, P, Lippke, J.A, Austen, D.A. | | Deposit date: | 2002-09-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Aurora-2, an Oncogenic Serine/Threonine Kinase
J.Biol.Chem., 277, 2002
|
|
4GDX
 
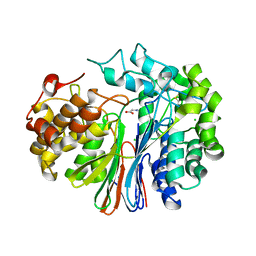 | | Crystal Structure of Human Gamma-Glutamyl Transpeptidase--Glutamate complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | West, M.B, Chen, Y, Wickham, S, Heroux, A, Cahill, K, Hanigan, M.H, Mooers, B.H.M. | | Deposit date: | 2012-08-01 | | Release date: | 2013-09-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Novel Insights into Eukaryotic gamma-Glutamyltranspeptidase 1 from the Crystal Structure of the Glutamate-bound Human Enzyme.
J.Biol.Chem., 288, 2013
|
|
5AOH
 
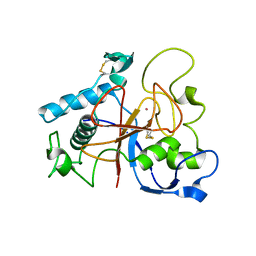 | | Crystal Structure of CarF | | Descriptor: | POTASSIUM ION, Spore coat protein CotH | | Authors: | Tichy, E.M, Hardwick, S.W, Luisi, B.F, C Salmond, G.P. | | Deposit date: | 2015-09-10 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 angstrom resolution crystal structure of the carbapenem intrinsic resistance protein CarF.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
2YJT
 
 | |
