5VNO
 
 | | Crystal structure of Sec23a/Sec24a/Sec22 | | Descriptor: | Protein transport protein Sec23A, Protein transport protein Sec24A, Vesicle-trafficking protein SEC22b, ... | | Authors: | Ma, W, Goldberg, J. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.905 Å) | | Cite: | ER retention is imposed by COPII protein sorting and attenuated by 4-phenylbutyrate.
Elife, 6, 2017
|
|
1LWV
 
 | | Borohydride-trapped hOgg1 Intermediate Structure Co-Crystallized with 8-aminoguanine | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(PED)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 8-AMINOGUANINE, ... | | Authors: | Fromme, J.C, Bruner, S.D, Yang, W, Karplus, M, Verdine, G.L. | | Deposit date: | 2002-06-03 | | Release date: | 2003-02-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Product-Assisted Catalysis in Base Excision DNA Repair
Nat.Struct.Biol., 10, 2003
|
|
5VPL
 
 | | CRYSTAL STRUCTURE OF DER F 1 COMPLEXED WITH FAB 4C1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4C1 - HEAVY CHAIN, ... | | Authors: | Chruszcz, M, Vailes, L.D, Chapman, M.D, Pomes, A, Minor, W. | | Deposit date: | 2017-05-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Determinants For Antibody Binding On Group 1 House Dust Mite Allergens.
J.Biol.Chem., 287, 2012
|
|
5VZY
 
 | | Crystal structure of crenezumab Fab in complex with Abeta | | Descriptor: | Amyloid beta A4 protein, Crenezumab Fab heavy chain,Immunoglobulin gamma-1 heavy chain, Crenezumab Fab light chain,Immunoblobulin light chain | | Authors: | Ultsch, M, Wang, W. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-09 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of Crenezumab Complex with Abeta Shows Loss of beta-Hairpin.
Sci Rep, 6, 2016
|
|
3QSL
 
 | | Structure of CAE31940 from Bordetella bronchiseptica RB50 | | Descriptor: | CITRIC ACID, Putative exported protein | | Authors: | Bajor, J, Kagan, O, Chruszcz, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-21 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of pyrimidine/thiamin biosynthesis precursor-like domain-containing protein CAE31940 from proteobacterium Bordetella bronchiseptica RB50, and evolutionary insight into the NMT1/THI5 family.
J Struct Funct Genomics, 15, 2014
|
|
1M0N
 
 | | Structure of Dialkylglycine Decarboxylase Complexed with 1-Aminocyclopentanephosphonate | | Descriptor: | 1-[((1E)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYLENE)AMINO]CYCLOPENTYLPHOSPHONIC ACID, 2,2-Dialkylglycine decarboxylase, POTASSIUM ION, ... | | Authors: | Liu, W, Rogers, C.J, Fisher, A.J, Toney, M.D. | | Deposit date: | 2002-06-13 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aminophosphonate Inhibitors of Dialkylglycine Decarboxylase: Structural Basis for Slow Binding Inhibition
Biochemistry, 41, 2002
|
|
3PZ1
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG) in Complex with BMS3 | | Descriptor: | (3R)-3-benzyl-4-[(4-methoxyphenyl)sulfonyl]-1-[(1-methyl-1H-imidazol-5-yl)methyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepine-7-carbonitrile, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guo, Z, Bon, R.S, Stigter, E.A, Waldmann, H, Alexandrov, K, Blankenfeldt, W, Goody, R.S. | | Deposit date: | 2010-12-14 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Guided Development of Selective RabGGTase Inhibitors.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
5ZHN
 
 | | Crystal structure of TrmD from Pseudomonas aeruginosa in complex with active-site inhibitor | | Descriptor: | N-({4-[(octylamino)methyl]phenyl}methyl)-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-5-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Pasunooti, K.K, Balamkundu, S, Wong, Y.W, Nah, Q, Liu, C.F, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Thienopyrimidinone Derivatives That Inhibit Bacterial tRNA (Guanine37-N1)-Methyltransferase (TrmD) by Restructuring the Active Site with a Tyrosine-Flipping Mechanism.
J.Med.Chem., 62, 2019
|
|
5Z6C
 
 | |
3PY8
 
 | | Crystal structure of a mutant of the large fragment of DNA polymerase I from thermus aquaticus in a closed ternary complex with DNA and ddCTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*A*AP*GP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Schnur, A, Marx, A, Welte, W, Diederichs, K. | | Deposit date: | 2010-12-12 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Learning from Directed Evolution: Thermus aquaticus DNA Polymerase Mutants with Translesion Synthesis Activity.
Chembiochem, 12, 2011
|
|
5W51
 
 | |
3PZ3
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG) in Complex with BMS-analogue 14 | | Descriptor: | 4-({(3R)-7-(5-formylfuran-2-yl)-4-[(4-methoxyphenyl)sulfonyl]-1-[(1-methyl-1H-imidazol-5-yl)methyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepin-3-yl}methyl)phenyl benzylcarbamate, CALCIUM ION, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Bon, R.S, Stigter, E.A, Waldmann, H, Alexandrov, K, Blankenfeldt, W, Goody, R.S. | | Deposit date: | 2010-12-14 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Development of Selective RabGGTase Inhibitors.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
5VNF
 
 | |
4NA0
 
 | | Crystal structure of mouse poly(ADP-ribose) glycohydrolase (PARG) catalytic domain with ADPRibose | | Descriptor: | IODIDE ION, Poly(ADP-ribose) glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Wang, Z, Cheng, Z, Xu, W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-29 | | Last modified: | 2014-09-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic and biochemical analysis of the mouse poly(ADP-ribose) glycohydrolase.
Plos One, 9, 2014
|
|
5Z9Z
 
 | | The C-terminal RRM domain of Arabidopsis SMALL RNA DEGRADING NUCLEASE 1 (E329A/E330A/E332A) | | Descriptor: | CITRATE ANION, Small RNA degrading nuclease 1 | | Authors: | Chen, J, Liu, L, You, C, Gu, J, Ruan, W, Zhang, L, Cao, C, Gan, J, Huang, Y, Chen, X, Ma, J. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Structural and biochemical insights into small RNA 3' end trimming by Arabidopsis SDN1.
Nat Commun, 9, 2018
|
|
5VRQ
 
 | | Crystal structure of Legionella pneumophila effector AnkC | | Descriptor: | Ankyrin repeat-containing protein | | Authors: | Kozlov, G, Wong, K, Wang, W, Skubak, P, Munoz-Escobar, J, Liu, Y, Pannu, N.S, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2017-05-11 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Ankyrin repeats as a dimerization module.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
5ZMR
 
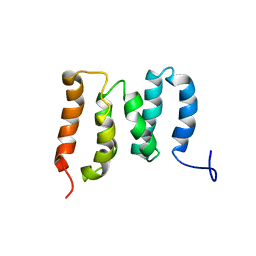 | | Solution Structure of the N-terminal Domain of the Yeast Rpn5 | | Descriptor: | 26S proteasome regulatory subunit RPN5 | | Authors: | Zhang, W, Zhao, C, Li, H, Hu, Y, Jin, C. | | Deposit date: | 2018-04-05 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of proteasome lid subunit Rpn5
Biochem. Biophys. Res. Commun., 504, 2018
|
|
1LWY
 
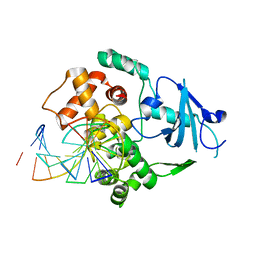 | | hOgg1 Borohydride-Trapped Intermediate without 8-oxoguanine | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(PED)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 8-OXOGUANINE DNA GLYCOSYLASE | | Authors: | Fromme, J.C, Bruner, S.D, Yang, W, Karplus, M, Verdine, G.L. | | Deposit date: | 2002-06-03 | | Release date: | 2003-02-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Product-Assisted Catalysis in Base Excision DNA Repair
Nat.Struct.Biol., 10, 2003
|
|
4NW2
 
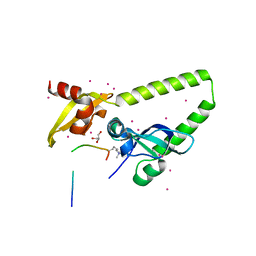 | | Tandem chromodomains of human CHD1 in complex with Influenza virus NS1 C-terminal tail trimethylated at K229 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, GLYCEROL, Nonstructural protein 1, ... | | Authors: | Qin, S, Tempel, W, Xu, C, El Bakkouri, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-05 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for histone mimicry and hijacking of host proteins by influenza virus protein NS1.
Nat Commun, 5, 2014
|
|
4AG9
 
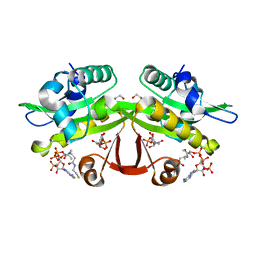 | | C. elegans glucosamine-6-phosphate N-acetyltransferase (GNA1): ternary complex with coenzyme A and GlcNAc | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, COENZYME A, ... | | Authors: | Dorfmueller, H.C, Fang, W, Rao, F.V, Blair, D.E, Attrill, H, Shepherd, S.M, van Aalten, D.M.F. | | Deposit date: | 2012-01-24 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and Biochemical Characterization of a Trapped Coenzyme a Adduct of Caenorhabditis Elegans Glucosamine-6-Phosphate N-Acetyltransferase 1.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2D55
 
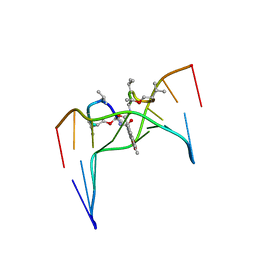 | | Structural, physical and biological characteristics of RNA.DNA binding agent N8-actinomycin D | | Descriptor: | ACTINOMYCIN D, DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*C)-3') | | Authors: | Shinomiya, M, Chu, W, Carlson, R.G, Weaver, R.F, Takusagawa, F. | | Deposit date: | 1995-05-01 | | Release date: | 1995-10-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the 2:1 Complex between D(Gaagcttc) and the Anticancer Drug Actinomycin D.
J.Mol.Biol., 225, 1992
|
|
4O1C
 
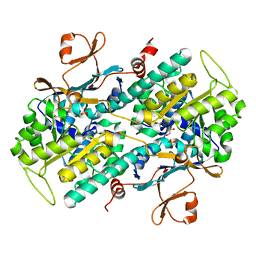 | | The crystal structures of a mutant NAMPT H191R | | Descriptor: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4AOA
 
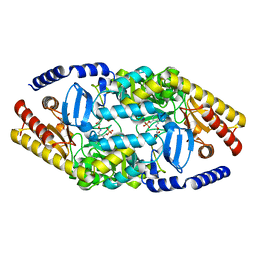 | | Biochemical properties and crystal structure of a novel beta- phenylalanine aminotransferase from Variovorax paradoxus | | Descriptor: | 4'-DEOXY-4'-ACETYLYAMINO-PYRIDOXAL-5'-PHOSPHATE, BETA-PHENYLALANINE AMINOTRANSFERASE, GLYCEROL | | Authors: | Crismaru, C.G, Wybenga, G.G, Szymanski, W, Wijma, H.J, Wu, B, deWildeman, S, Poelarends, G.J, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2012-03-25 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Biochemical Properties and Crystal Structure of a Beta-Phenylalanine Aminotransferase from Variovorax Paradoxus.
Appl.Environ.Microbiol., 79, 2013
|
|
5W5E
 
 | | Re-refinement of the pyocin tube structure | | Descriptor: | FIIR2 protein | | Authors: | Wang, F, Zheng, W, Taylor, N.M, Guerrero-Ferreira, R.C, Leiman, P.G, Egelman, E.H. | | Deposit date: | 2017-06-15 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Refined Cryo-EM Structure of the T4 Tail Tube: Exploring the Lowest Dose Limit.
Structure, 25, 2017
|
|
4NC3
 
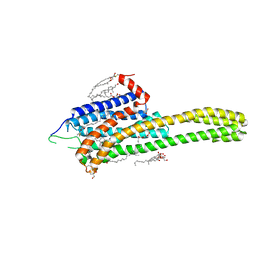 | | Crystal structure of the 5-HT2B receptor solved using serial femtosecond crystallography in lipidic cubic phase. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHOLESTEROL, ... | | Authors: | Liu, W, Wacker, D, Gati, C, Han, G.W, James, D, Wang, D, Nelson, G, Weierstall, U, Katritch, V, Barty, A, Zatsepin, N.A, Li, D, Messerschmidt, M, Boutet, S, Williams, G.J, Koglin, J.E, Seibert, M.M, Wang, C, Shah, S.T.A, Basu, S, Fromme, R, Kupitz, C, Rendek, K.N, Grotjohann, I, Fromme, P, Kirian, R.A, Beyerlein, K.R, White, T.A, Chapman, H.N, Caffrey, M, Spence, J.C.H, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Serial femtosecond crystallography of G protein-coupled receptors.
Science, 342, 2013
|
|
