2A3S
 
 | |
2D2W
 
 | |
2KIU
 
 | |
6CIM
 
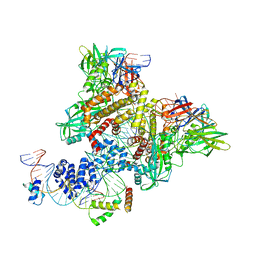 | | Pre-Reaction Complex, RAG1(E962Q)/2-nicked/intact 12/23RSS complex in Mn2+ | | Descriptor: | DNA (5'-D(*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3'), High mobility group protein B1, Intact 23RSS substrate forward strand, ... | | Authors: | Chuenchor, W, Chen, X, Kim, M.S, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
2MS9
 
 | | Solution structure of a G-quadruplex | | Descriptor: | DNA (28-MER) | | Authors: | Chung, W.J, Heddi, B, Schmitt, E, Lim, K.W, Mechulam, Y, Phan, A.T. | | Deposit date: | 2014-07-25 | | Release date: | 2015-02-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of a left-handed DNA G-quadruplex
Proc.Natl.Acad.Sci.USA, 2015
|
|
6OFC
 
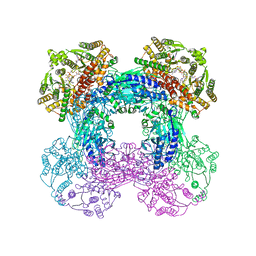 | | Crystal structure of M. tuberculosis glutamine-dependent NAD+ synthetase complexed with Sulfonamide derivative 1, pyrophosphate, and glutamine | | Descriptor: | 5'-O-[(pyridine-3-carbonyl)sulfamoyl]adenosine, CHLORIDE ION, GLUTAMINE, ... | | Authors: | Chuenchor, W, Doukov, T.I, Gerratana, B. | | Deposit date: | 2019-03-28 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Different ways to transport ammonia in human and Mycobacterium tuberculosis NAD+synthetases.
Nat Commun, 11, 2020
|
|
6CIL
 
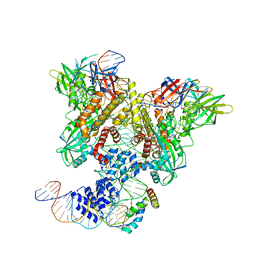 | | PRE-REACTION COMPLEX, RAG1(E962Q)/2-INTACT/INTACT 12/23RSS COMPLEX IN MN2+ | | Descriptor: | High mobility group protein B1, Intact 12RSS substrate forward strand, Intact 12RSS substrate reverse strand, ... | | Authors: | Chuenchor, W, Chen, X, Kim, M.S, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
6CIK
 
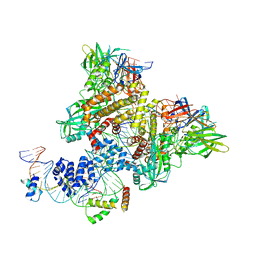 | | Pre-Reaction Complex, RAG1(E962Q)/2-intact/nicked 12/23RSS complex in Mn2+ | | Descriptor: | DNA (5'-D(*AP*TP*CP*TP*GP*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3'), High mobility group protein B1, Intact 12RSS substrate forward strand, ... | | Authors: | Chuenchor, W, Chen, X, Kim, M.S, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
1TRQ
 
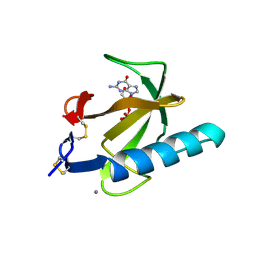 | |
1A6B
 
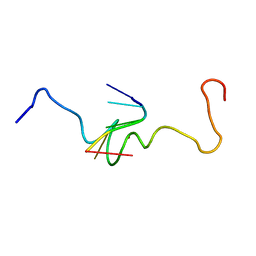 | | NMR STRUCTURE OF THE COMPLEX BETWEEN THE ZINC FINGER PROTEIN NCP10 OF MOLONEY MURINE LEUKEMIA VIRUS AND A SEQUENCE OF THE PSI-PACKAGING DOMAIN OF HIV-1, 20 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*C)-3'), ZINC FINGER PROTEIN NCP10, ZINC ION | | Authors: | Schueler, W, Dong, C.-Z, Wecker, K, Roques, B.P. | | Deposit date: | 1998-02-23 | | Release date: | 1999-08-23 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the complex between the zinc finger protein NCp10 of Moloney murine leukemia virus and the single-stranded pentanucleotide d(ACGCC): comparison with HIV-NCp7 complexes.
Biochemistry, 38, 1999
|
|
2JTC
 
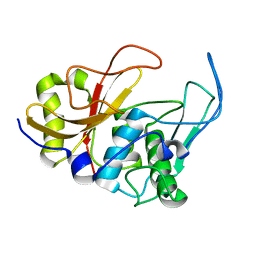 | | 3D structure and backbone dynamics of SPE B | | Descriptor: | Streptopain | | Authors: | Chuang, W, Wang, C, Houng, H, Chen, C, Wang, P. | | Deposit date: | 2007-07-26 | | Release date: | 2008-08-26 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of streptopain: insight into diverse substrate specificity.
J.Biol.Chem., 284, 2009
|
|
1JXS
 
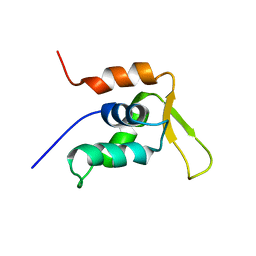 | | Solution Structure of the DNA-Binding Domain of Interleukin Enhancer Binding Factor | | Descriptor: | interleukin enhancer binding factor | | Authors: | Chuang, W.J, Liu, P.P, Li, C, Hsieh, Y.H, Chen, S.W, Chen, S.H, Jeng, W.Y. | | Deposit date: | 2001-09-08 | | Release date: | 2003-03-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of interleukin enhancer binding factor 1 (FOXK1a)
PROTEINS: STRUCT.,FUNCT.,GENET., 49, 2002
|
|
1H6T
 
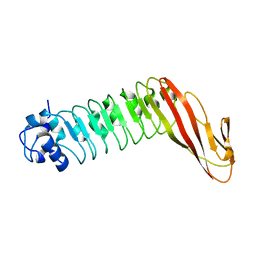 | | Internalin B: crystal structure of fused N-terminal domains. | | Descriptor: | INTERNALIN B | | Authors: | Schubert, W.-D, Gobel, G, Diepholz, M, Darji, A, Kloer, D, Hain, T, Chakraborty, T, Wehland, J, Domann, E, Heinz, D.W. | | Deposit date: | 2001-06-22 | | Release date: | 2001-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Internalins from the human pathogen Listeria monocytogenes combine three distinct folds into a contiguous internalin domain.
J.Mol.Biol., 312, 2001
|
|
2RGL
 
 | | Rice BGlu1 beta-glucosidase, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2007-10-04 | | Release date: | 2008-02-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into Rice BGlu1 beta-Glucosidase Oligosaccharide Hydrolysis and Transglycosylation
J.Mol.Biol., 377, 2008
|
|
6OFB
 
 | | Crystal structure of human glutamine-dependent NAD+ synthetase complexed with NaAD+, AMP, pyrophosphate, and Mg2+ | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Glutamine-dependent NAD(+) synthetase, ... | | Authors: | Chuenchor, W, Doukov, T.I, Gerratana, B. | | Deposit date: | 2019-03-28 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Different ways to transport ammonia in human and Mycobacterium tuberculosis NAD+synthetases.
Nat Commun, 11, 2020
|
|
2RGM
 
 | | Rice BGlu1 beta-glucosidase, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-deoxy-2-fluoro-alpha-D-glucopyranose, Beta-glucosidase, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2007-10-04 | | Release date: | 2008-02-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Insights into Rice BGlu1 beta-Glucosidase Oligosaccharide Hydrolysis and Transglycosylation
J.Mol.Biol., 377, 2008
|
|
1VPC
 
 | | C-TERMINAL DOMAIN (52-96) OF THE HIV-1 REGULATORY PROTEIN VPR, NMR, 1 STRUCTURE | | Descriptor: | VPR PROTEIN | | Authors: | Schueler, W, De Rocquigny, H, Baudat, Y, Sire, J, Roques, B.P. | | Deposit date: | 1998-02-20 | | Release date: | 1999-03-23 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the (52-96) C-terminal domain of the HIV-1 regulatory protein Vpr: molecular insights into its biological functions.
J.Mol.Biol., 285, 1999
|
|
2PJF
 
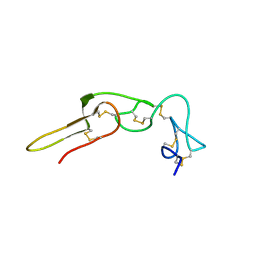 | | Solution structure of rhodostomin | | Descriptor: | Rhodostoxin-disintegrin rhodostomin | | Authors: | Chuang, W.J, Chen, Y.C, Chen, C.Y, Chang, Y.T. | | Deposit date: | 2007-04-16 | | Release date: | 2007-05-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Effect of D to E mutation of the RGD motif in rhodostomin on its activity, structure, and dynamics: Importance of the interactions between the D residue and integrin
Proteins, 2009
|
|
2PJG
 
 | | Solution structure of rhodostomin D51E mutant | | Descriptor: | Rhodostoxin-disintegrin rhodostomin | | Authors: | Chuang, W.J, Chen, Y.C, Chen, C.Y, Chou, L.J. | | Deposit date: | 2007-04-16 | | Release date: | 2007-05-08 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Effect of D to E mutation of the RGD motif in rhodostomin on its activity, structure, and dynamics: Importance of the interactions between the D residue and integrin
Proteins, 2009
|
|
1O6T
 
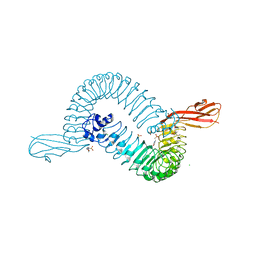 | | Internalin (INLA, Listeria monocytogenes) - functional domain, uncomplexed | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schubert, W.-D, Urbanke, C, Ziehm, T, Beier, V, Machner, M.P, Domann, E, Wehland, J, Chakraborty, T, Heinz, D.W. | | Deposit date: | 2002-10-15 | | Release date: | 2002-12-23 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Internalin, a Major Invasion Protein of Listeria Monocytogenes, in Complex with its Human Receptor E-Cadherin
Cell(Cambridge,Mass.), 111, 2002
|
|
1O6V
 
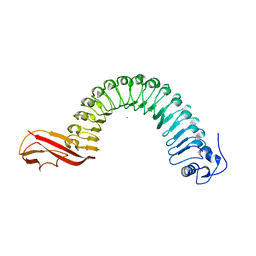 | | Internalin (INLA, Listeria monocytogenes) - functional domain, uncomplexed | | Descriptor: | CALCIUM ION, INTERNALIN A | | Authors: | Schubert, W.-D, Urbanke, C, Ziehm, T, Beier, V, Machner, M.P, Domann, E, Wehland, J, Chakraborty, T, Heinz, D.W. | | Deposit date: | 2002-10-16 | | Release date: | 2002-12-23 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Internalin, a Major Invasion Protein of Listeria Monocytogenes, in Complex with its Human Receptor E-Cadherin
Cell(Cambridge,Mass.), 111, 2002
|
|
4TTC
 
 | |
1O6S
 
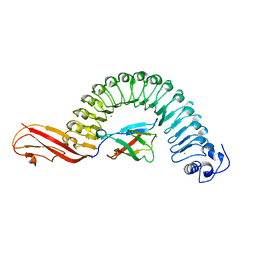 | | Internalin (Listeria monocytogenes) / E-Cadherin (human) Recognition Complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, E-CADHERIN, ... | | Authors: | Schubert, W.-D, Urbanke, C, Ziehm, T, Beier, V, Machner, M.P, Domann, E, Wehland, J, Chakraborty, T, Heinz, D.W. | | Deposit date: | 2002-10-13 | | Release date: | 2002-12-13 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Internalin, a Major Invasion Protein of Listeria Monocytogenes, in Complex with its Human Receptor E-Cadherin
Cell(Cambridge,Mass.), 111, 2002
|
|
6UP2
 
 | |
6UP3
 
 | |
