6INZ
 
 | | Crystal structure of solute-binding protein complexed with unsaturated hyaluronan disaccharide | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular solute-binding protein family 1, ... | | Authors: | Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2018-10-29 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Substrate recognition by bacterial solute-binding protein is responsible for import of extracellular hyaluronan and chondroitin sulfate from the animal host.
Biosci.Biotechnol.Biochem., 83, 2019
|
|
5GUB
 
 | | Crystal structure of solute-binding protein complexed with sulfate group-free unsaturated chondroitin disaccharide | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2016-08-26 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A bacterial ABC transporter enables import of mammalian host glycosaminoglycans
Sci Rep, 7, 2017
|
|
5GX8
 
 | | Crystal structure of solute-binding protein related to glycosaminoglycans | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, D(-)-TARTARIC ACID, ... | | Authors: | Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2016-09-15 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.778 Å) | | Cite: | A bacterial ABC transporter enables import of mammalian host glycosaminoglycans
Sci Rep, 7, 2017
|
|
5GX6
 
 | | Crystal structure of solute-binding protein complexed with unsaturated chondroitin disaccharide with a sulfate group at C-4 position of GalNAc | | Descriptor: | 1,2-ETHANEDIOL, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2016-09-15 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A bacterial ABC transporter enables import of mammalian host glycosaminoglycans
Sci Rep, 7, 2017
|
|
5GX7
 
 | | Crystal structure of solute-binding protein complexed with unsaturated chondroitin disaccharide with a sulfate group at C-6 position of GalNAc | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-6-O-sulfo-beta-D-galactopyranose, CALCIUM ION, Extracellular solute-binding protein family 1 | | Authors: | Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2016-09-15 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | A bacterial ABC transporter enables import of mammalian host glycosaminoglycans
Sci Rep, 7, 2017
|
|
5XS8
 
 | | Crystal structure of solute-binding protein complexed with unsaturated chondroitin disaccharide with two sulfate groups at C-4 and C-6 positions of GalNAc | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4,6-di-O-sulfo-beta-D-galactopyranose, CALCIUM ION, Extracellular solute-binding protein family 1 | | Authors: | Oiki, S, Kamochi, R, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2017-06-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Alternative substrate-bound conformation of bacterial solute-binding protein involved in the import of mammalian host glycosaminoglycans.
Sci Rep, 7, 2017
|
|
3WUX
 
 | | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N/K370S from Streptococcus agalactiae | | Descriptor: | 1,2-ETHANEDIOL, Unsaturated chondroitin disaccharide hydrolase | | Authors: | Nakamichi, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-05-08 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N/K370S from Streptococcus agalactiae
to be published
|
|
4U8E
 
 | | Crystal structure of 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase from Streptococcus agalactiae | | Descriptor: | Putative uncharacterized protein gbs1892 | | Authors: | Maruyama, y, Oiki, S, Takase, R, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-03 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metabolic Fate of Unsaturated Glucuronic/Iduronic Acids from Glycosaminoglycans: MOLECULAR IDENTIFICATION AND STRUCTURE DETERMINATION OF STREPTOCOCCAL ISOMERASE AND DEHYDROGENASE
J.Biol.Chem., 290, 2015
|
|
4TKZ
 
 | | Crystal structure of phosphotransferase system component EIIA from Streptococcus agalactiae | | Descriptor: | GLYCEROL, Putative uncharacterized protein gbs1890 | | Authors: | Nakamichi, Y, Maruyama, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-05-28 | | Release date: | 2014-08-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of phosphotransferase system component EIIA from Streptococcus agalactiae
To Be Published
|
|
4Z9Y
 
 | | Crystal structure of 2-keto-3-deoxy-D-gluconate dehydrogenase from Pectobacterium carotovorum | | Descriptor: | 2-deoxy-D-gluconate 3-dehydrogenase, SULFATE ION | | Authors: | Takase, R, Maruyama, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural determinants in bacterial 2-keto-3-deoxy-D-gluconate dehydrogenase KduD for dual-coenzyme specificity
Proteins, 84, 2016
|
|
4Z9X
 
 | | Crystal structure of 2-keto-3-deoxy-D-gluconate dehydrogenase from Streptococcus pyogenes | | Descriptor: | Gluconate 5-dehydrogenase | | Authors: | Maruyama, Y, Takase, R, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural determinants in bacterial 2-keto-3-deoxy-D-gluconate dehydrogenase KduD for dual-coenzyme specificity
Proteins, 84, 2016
|
|
4ZA2
 
 | | Crystal structure of Pectobacterium carotovorum 2-keto-3-deoxy-D-gluconate dehydrogenase complexed with NAD+ | | Descriptor: | 2-deoxy-D-gluconate 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Takase, R, Maruyama, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural determinants in bacterial 2-keto-3-deoxy-D-gluconate dehydrogenase KduD for dual-coenzyme specificity
Proteins, 84, 2016
|
|
7YE3
 
 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase, ZINC ION | | Authors: | Yamamoto, Y, Oiki, S, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Crystal Structures of Lacticaseibacillus 4-Deoxy-L- threo- 5-hexosulose-uronate Ketol-isomerase KduI in Complex with Substrate Analogs.
J Appl Glycosci (1999), 70, 2023
|
|
4U8F
 
 | | Crystal structure of 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase complexed with a tartrate | | Descriptor: | L(+)-TARTARIC ACID, Putative uncharacterized protein gbs1892 | | Authors: | Maruyama, Y, Oiki, S, Takase, R, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Metabolic Fate of Unsaturated Glucuronic/Iduronic Acids from Glycosaminoglycans: MOLECULAR IDENTIFICATION AND STRUCTURE DETERMINATION OF STREPTOCOCCAL ISOMERASE AND DEHYDROGENASE.
J.Biol.Chem., 290, 2015
|
|
4U8G
 
 | | Crystal structure of 2-keto-3-deoxy-D-gluconate dehydrogenase from Streptococcus agalactiae | | Descriptor: | Putative uncharacterized protein gbs1891 | | Authors: | Maruyama, Y, Oiki, S, Takase, R, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Metabolic Fate of Unsaturated Glucuronic/Iduronic Acids from Glycosaminoglycans: MOLECULAR IDENTIFICATION AND STRUCTURE DETERMINATION OF STREPTOCOCCAL ISOMERASE AND DEHYDROGENASE.
J.Biol.Chem., 290, 2015
|
|
7YRS
 
 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with MOPS | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase, ZINC ION | | Authors: | Yamamoto, Y, Oiki, S, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2022-08-10 | | Release date: | 2023-08-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Crystal Structures of Lacticaseibacillus 4-Deoxy-L- threo- 5-hexosulose-uronate Ketol-isomerase KduI in Complex with Substrate Analogs.
J Appl Glycosci (1999), 70, 2023
|
|
7VGK
 
 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI | | Descriptor: | 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase | | Authors: | Iwase, H, Oiki, S, Mikami, B, Takase, R, Hashimoto, W. | | Deposit date: | 2021-09-16 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of Lacticaseibacillus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI in complex with substrate analogs
J.Appl.Glyosci., 2023
|
|
5Z6B
 
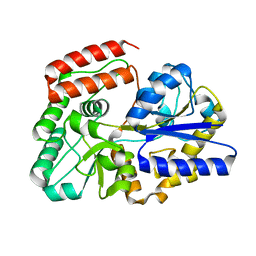 | | Crystal structure of sugar-binding protein YesO in complex with rhamnogalacturonan trisaccharide | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-[beta-D-galactopyranose-(1-4)]alpha-L-rhamnopyranose, Putative ABC transporter substrate-binding protein YesO | | Authors: | Sugiura, H, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2018-01-22 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.582 Å) | | Cite: | Crystal structure of sugar-binding protein YesO in complex with rhamnogalacturonan trisaccharide
To Be Published
|
|
5Z6C
 
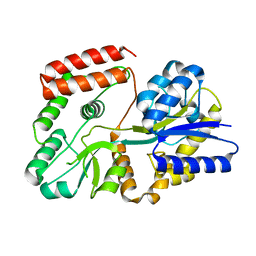 | |
