8ANE
 
 | | Structure of the type I-G CRISPR effector | | Descriptor: | Cas7, RNA (66-MER) | | Authors: | Shangguan, Q, Graham, S, Sundaramoorthy, R, White, M.F. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and mechanism of the type I-G CRISPR effector.
Nucleic Acids Res., 50, 2022
|
|
1FBZ
 
 | | Structure-based design of a novel, osteoclast-selective, nonpeptide Src SH2 inhibitor with in vivo anti-resorptive activity | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE LCK, {4-[2-ACETYLAMINO-2-(3-CARBAMOYL-2-CYCLOHEXYLMETHOXY-6,7,8,9-TETRAHYDRO-5H-BENZOCYCLOHEPTEN-5YLCARBAMOYL)-ETHYL]-2-PHOSPHONO-PHENYL}-PHOSPHONIC ACID | | Authors: | Shakespeare, W, Yang, M, Bohacek, R, Cerasoli, F, Stebbis, K, Sundaramoorthi, R, Vu, C, Pradeepan, S, Metcalf, C, Haraldson, C, Merry, T, Dalgarno, D, Narula, S, Hatada, M, Lu, X, Van Schravendijk, M.R, Adams, S, Violette, S, Smith, J, Guan, W, Bartlett, C, Herson, J, Iuliucci, J, Weigele, M, Sawyer, T. | | Deposit date: | 2000-07-17 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of an osteoclast-selective, nonpeptide src homology 2 inhibitor with in vivo antiresorptive activity.
Proc.Natl.Acad.Sci.Usa, 97, 2000
|
|
5N2W
 
 | | WT-Parkin and pUB complex | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin,E3 ubiquitin-protein ligase parkin, Polyubiquitin-B, ... | | Authors: | Kumar, A, Chaugule, V.K, Johnson, C, Toth, R, Sundaramoorthy, R, Knebel, A, Walden, H. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5N38
 
 | | S65DParkin and pUB complex | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase parkin,E3 ubiquitin-protein ligase parkin, ... | | Authors: | Kumar, A, Chaugule, V.K, Johnson, C, Toth, R, Sundaramoorthy, R, Knebel, A, Walden, H. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity.
Nat. Struct. Mol. Biol., 24, 2017
|
|
4A7Z
 
 | | Complex of bifunctional aldos-2-ulose dehydratase with the reaction intermediate ascopyrone M | | Descriptor: | ALDOS-2-ULOSE DEHYDRATASE, Ascopyrone M, MAGNESIUM ION, ... | | Authors: | Claesson, M, Lindqvist, Y, Madrid, S, Sandalova, T, Fiskesund, R, Yu, S, Schneider, G. | | Deposit date: | 2011-11-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Bifunctional Aldos-2-Ulose Dehydratase/Isomerase from Phanerochaete Chrysosporium with the Reaction Intermediate Ascopyrone M.
J.Mol.Biol., 417, 2012
|
|
4A7Y
 
 | | Active site metal depleted aldos-2-ulose dehydratase | | Descriptor: | 1,5-anhydro-D-fructose, ALDOS-2-ULOSE DEHYDRATASE, MAGNESIUM ION, ... | | Authors: | Claesson, M, Lindqvist, Y, Madrid, S, Sandalova, T, Fiskesund, R, Yu, S, Schneider, G. | | Deposit date: | 2011-11-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Bifunctional Aldos-2-Ulose Dehydratase/Isomerase from Phanerochaete Chrysosporium with the Reaction Intermediate Ascopyrone M.
J.Mol.Biol., 417, 2012
|
|
4A7K
 
 | | Bifunctional Aldos-2-ulose dehydratase | | Descriptor: | ALDOS-2-ULOSE DEHYDRATASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Claesson, M, Lindqvist, Y, Madrid, S, Sandalova, T, Fiskesund, R, Yu, S, Schneider, G. | | Deposit date: | 2011-11-14 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Bifunctional Aldos-2-Ulose Dehydratase/Isomerase from Phanerochaete Chrysosporium with the Reaction Intermediate Ascopyrone M.
J.Mol.Biol., 417, 2012
|
|
8B2X
 
 | |
1V5C
 
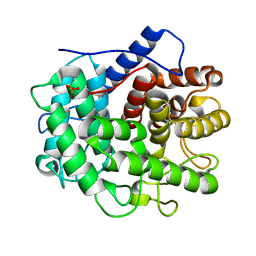 | | The crystal structure of the inactive form chitosanase from Bacillus sp. K17 at pH3.7 | | Descriptor: | SULFATE ION, chitosanase | | Authors: | Adachi, W, Shimizu, S, Sunami, T, Fukazawa, T, Suzuki, M, Yatsunami, R, Nakamura, S, Takenaka, A. | | Deposit date: | 2003-11-22 | | Release date: | 2004-12-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of family GH-8 chitosanase with subclass II specificity from Bacillus sp. K17
J.MOL.BIOL., 343, 2004
|
|
1V5D
 
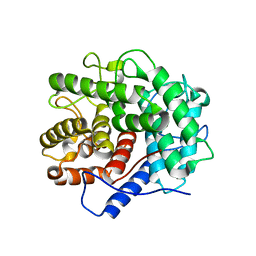 | | The crystal structure of the active form chitosanase from Bacillus sp. K17 at pH6.4 | | Descriptor: | PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), chitosanase | | Authors: | Adachi, W, Shimizu, S, Sunami, T, Fukazawa, T, Suzuki, M, Yatsunami, R, Nakamura, S, Takenaka, A. | | Deposit date: | 2003-11-22 | | Release date: | 2004-12-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of family GH-8 chitosanase with subclass II specificity from Bacillus sp. K17
J.MOL.BIOL., 343, 2004
|
|
8OV6
 
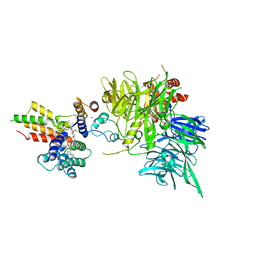 | | Ternary structure of intramolecular bivalent glue degrader IBG1 bound to BRD4 and DCAF16:DDB1deltaBPB | | Descriptor: | Bromodomain-containing protein 4, DDB1- and CUL4-associated factor 16, DDB1deltaBPB, ... | | Authors: | Cowan, A.D, Sundaramoorthy, R, Nakasone, M.A, Ciulli, A. | | Deposit date: | 2023-04-25 | | Release date: | 2023-05-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Targeted protein degradation via intramolecular bivalent glues.
Nature, 627, 2024
|
|
1CS4
 
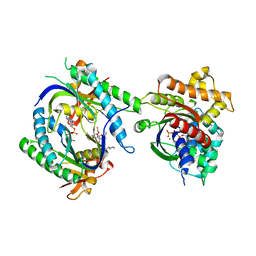 | | COMPLEX OF GS-ALPHA WITH THE CATALYTIC DOMAINS OF MAMMALIAN ADENYLYL CYCLASE: COMPLEX WITH 2'-DEOXY-ADENOSINE 3'-MONOPHOSPHATE, PYROPHOSPHATE AND MG | | Descriptor: | 2'-DEOXY-ADENOSINE 3'-MONOPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Tesmer, J.J.G, Dessauer, C.A, Sunahara, R.K, Johnson, R.A, Gilman, A.G, Sprang, S.R. | | Deposit date: | 1999-08-16 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for P-site inhibition of adenylyl cyclase.
Biochemistry, 39, 2000
|
|
3PDS
 
 | | Irreversible Agonist-Beta2 Adrenoceptor Complex | | Descriptor: | 8-hydroxy-5-[(1R)-1-hydroxy-2-({2-[3-methoxy-4-(3-sulfanylpropoxy)phenyl]ethyl}amino)ethyl]quinolin-2(1H)-one, CHOLESTEROL, Fusion protein Beta-2 adrenergic receptor/Lysozyme, ... | | Authors: | Rosenbaum, D.M, Zhang, C, Lyons, J.A, Holl, R, Aragao, D, Arlow, D.H, Rasmussen, S.G.F, Choi, H.-J, DeVree, B.T, Sunahara, R.K, Chae, P.S, Gellman, S.H, Dror, R.O, Shaw, D.E, Weis, W.I, Caffrey, M, Gmeiner, P, Kobilka, B.K. | | Deposit date: | 2010-10-24 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and function of an irreversible agonist-beta(2) adrenoceptor complex
Nature, 469, 2011
|
|
1CUL
 
 | | COMPLEX OF GS-ALPHA WITH THE CATALYTIC DOMAINS OF MAMMALIAN ADENYLYL CYCLASE: COMPLEX WITH 2',5'-DIDEOXY-ADENOSINE 3'-TRIPHOSPHATE AND MG | | Descriptor: | 2',5'-DIDEOXY-ADENOSINE 3'-MONOPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Tesmer, J.J.G, Dessauer, C.A, Sunahara, R.K, Johnson, R.A, Gilman, A.G, Sprang, S.R. | | Deposit date: | 1999-08-20 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for P-site inhibition of adenylyl cyclase.
Biochemistry, 39, 2000
|
|
5AGC
 
 | | Crystallographic forms of the Vps75 tetramer | | Descriptor: | VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 75 | | Authors: | Hammond, C.M, Sundaramoorthy, R, Owen-Hughes, T. | | Deposit date: | 2015-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Histone Chaperone Vps75 Forms Multiple Oligomeric Assemblies Capable of Mediating Exchange between Histone H3-H4 Tetramers and Asf1-H3-H4 Complexes.
Nucleic Acids Res., 44, 2016
|
|
6MXT
 
 | | Crystal structure of human beta2 adrenergic receptor bound to salmeterol and Nb71 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Endolysin, ... | | Authors: | Masureel, M, Zou, Y, Picard, L.P, van der Westhuizen, E, Mahoney, J.P, Rodrigues, J.P.G.L.M, Mildorf, T.J, Dror, R.O, Shaw, D.E, Bouvier, M, Pardon, E, Steyaert, J, Sunahara, R.K, Weis, W.I, Zhang, C, Kobilka, B.K. | | Deposit date: | 2018-10-31 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95934224 Å) | | Cite: | Structural insights into binding specificity, efficacy and bias of a beta2AR partial agonist.
Nat. Chem. Biol., 14, 2018
|
|
6OBA
 
 | | The beta2 adrenergic receptor bound to a negative allosteric modulator | | Descriptor: | (2S)-1-[(1-methylethyl)amino]-3-(2-prop-2-en-1-ylphenoxy)propan-2-ol, 6-bromo-N~2~-phenylquinazoline-2,4-diamine, Beta-2 adrenergic receptor,Lysozyme,Beta-2 adrenergic receptor, ... | | Authors: | Liu, X, Stobel, A, Kaindl, J, Dengler, D, ClarK, M, Mahoney, J, Korczynska, M, Matt, R.A, Hubner, H, Xu, X, Stanek, M, Hirata, K, Shoichet, B, Sunahara, R, Gmeiner, R, Kobilka, B.K. | | Deposit date: | 2019-03-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An allosteric modulator binds to a conformational hub in the beta2adrenergic receptor.
Nat.Chem.Biol., 16, 2020
|
|
4RMB
 
 | |
5C23
 
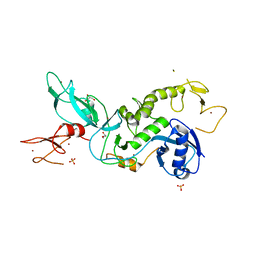 | | Parkin (S65DUblR0RBR) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | Kumar, A, Aguirre, J.D, Condos, T.E.C, Martinez-Torres, R.J, Chaugule, V.K, Toth, R, Sundaramoorthy, R, Mercier, P, Knebel, A, Spratt, D.E, Barber, K.R, Shaw, G.S, Walden, H. | | Deposit date: | 2015-06-15 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis.
Embo J., 34, 2015
|
|
5C1Z
 
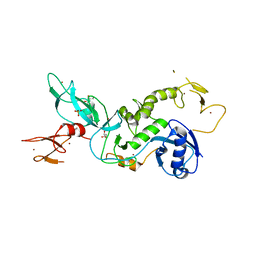 | | Parkin (UblR0RBR) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | kumar, A, Aguirre, J.D, Condos, T.E.C, Martinez-Torres, R.J, Chaugule, V.K, Toth, R, Sundaramoorthy, R, Mercier, P, Knebel, A, Spratt, D.E, Barber, K.R, Shaw, G.S, Walden, H. | | Deposit date: | 2015-06-15 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis.
Embo J., 34, 2015
|
|
4DKL
 
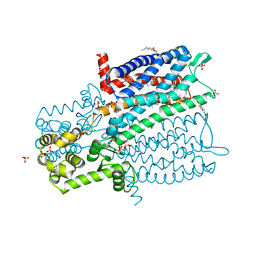 | | Crystal structure of the mu-opioid receptor bound to a morphinan antagonist | | Descriptor: | CHLORIDE ION, CHOLESTEROL, Mu-type opioid receptor, ... | | Authors: | Manglik, A, Kruse, A.C, Kobilka, T.S, Thian, F.S, Mathiesen, J.M, Sunahara, R.K, Pardo, L, Weis, W.I, Kobilka, B.K, Granier, S. | | Deposit date: | 2012-02-03 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the {mu}-opioid receptor bound to a morphinan antagonist.
Nature, 485, 2012
|
|
6VMS
 
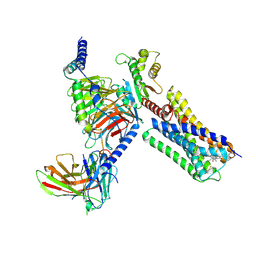 | | Structure of a D2 dopamine receptor-G-protein complex in a lipid membrane | | Descriptor: | Endolysin,D(2) dopamine receptor,D(2) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yin, J, Chen, K.M, Clark, M.J, Hijazi, M, Kumari, P, Bai, X, Sunahara, R.K, Barth, P, Rosenbaum, D.M. | | Deposit date: | 2020-01-28 | | Release date: | 2020-06-17 | | Last modified: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a D2 dopamine receptor-G-protein complex in a lipid membrane.
Nature, 584, 2020
|
|
7XK9
 
 | | Structure of human beta2 adrenergic receptor bound to constrained isoproterenol | | Descriptor: | (5R,6R)-6-(propan-2-ylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Camelid Antibody Fragment, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Xu, X, Shonberg, J, Kaindl, J, Clark, M, Stobel, A, Maul, L, Mayer, D, Hubner, H, Venkatakrishnan, A, Dror, R, Kobilka, B.K, Sunahara, R, Liu, X, Gmeiner, P. | | Deposit date: | 2022-04-19 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Constrained catecholamines gain beta 2 AR selectivity through allosteric effects on pocket dynamics.
Nat Commun, 14, 2023
|
|
7XKA
 
 | | Structure of human beta2 adrenergic receptor bound to constrained epinephrine | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Camelid Antibody Fragment, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Xu, X, Shonberg, J, Kaindl, J, Clark, M, Stobel, A, Maul, L, Mayer, D, Hubner, H, Venkatakrishnan, A, Dror, R, Kobilka, B.K, Sunahara, R, Liu, X, Gmeiner, P. | | Deposit date: | 2022-04-19 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Constrained catecholamines gain beta 2 AR selectivity through allosteric effects on pocket dynamics.
Nat Commun, 14, 2023
|
|
2XB0
 
 | |
