4LQ6
 
 | | Crystal structure of Rv3717 reveals a novel amidase from M. tuberculosis | | Descriptor: | CHLORIDE ION, N-acetymuramyl-L-alanine amidase-related protein, PLATINUM (II) ION, ... | | Authors: | Kumar, A, Kumar, S, Kumar, D, Mishra, A, Dewangan, R.P, Shrivastava, P, Ramachandran, S, Taneja, B. | | Deposit date: | 2013-07-17 | | Release date: | 2013-12-04 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The structure of Rv3717 reveals a novel amidase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3KJZ
 
 | | Crystal structure of native peptidyl-tRNA hydrolase from Mycobacterium smegmatis | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Kumar, A, Singh, N, Yadav, R, Prem Kumar, R, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2009-11-04 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from mycobacterium smegmatis reveals novel features related to enzyme dynamics.
Int J Biochem Mol Biol, 3, 2012
|
|
5FCF
 
 | | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris, phosphate and Mn bound | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLY-GLY-GLY, GLYCEROL, ... | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R.D. | | Deposit date: | 2015-12-15 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and biochemical investigations reveal novel mode of substrate selectivity and illuminate substrate inhibition and allostericity in a subfamily of Xaa-Pro dipeptidases.
Biochim. Biophys. Acta, 1865, 2017
|
|
5FCH
 
 | | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris, phosphate and Zn bound | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLY-GLY-GLY, GLYCEROL, ... | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R.D. | | Deposit date: | 2015-12-15 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and biochemical investigations reveal novel mode of substrate selectivity and illuminate substrate inhibition and allostericity in a subfamily of Xaa-Pro dipeptidases
Biochim. Biophys. Acta, 1865, 2017
|
|
8BW0
 
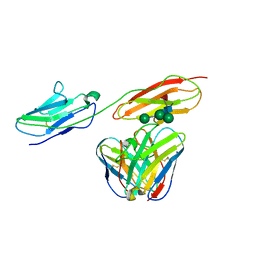 | | Structure of CEACAM5 A3-B3 domain in Complex with Tusamitamab Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Carcinoembryonic antigen-related cell adhesion molecule 5, ... | | Authors: | Kumar, A, Bertrand, T, Rapisarda, C, Rak, A. | | Deposit date: | 2022-12-06 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural insights into epitope-paratope interactions of monoclonal antibody targeting CEACAM5-expressing tumors
Res Sq, 2023
|
|
3KK0
 
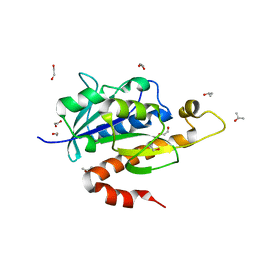 | | Crystal structure of partially folded intermediate state of peptidyl-tRNA hydrolase from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, A, Singh, N, Yadav, R, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2009-11-04 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of Fully-Folded Native and Partially-Folded Intermediate States of Peptidyl-tRNA Hydrolase from Mycobacterium smegmatis
To be Published
|
|
6FC5
 
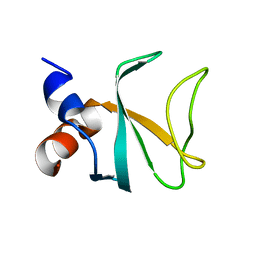 | | Bik1 CAP-Gly domain | | Descriptor: | Microtubule-associated protein | | Authors: | Kumar, A, Stangier, M.M, Steinmetz, M.O. | | Deposit date: | 2017-12-20 | | Release date: | 2018-04-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure-Function Relationship of the Bik1-Bim1 Complex.
Structure, 26, 2018
|
|
6VM2
 
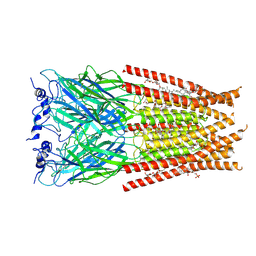 | | Full length Glycine receptor reconstituted in lipid nanodisc in Gly/IVM-conformation (State-2) | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kumar, A, Basak, S, Chakrapani, S. | | Deposit date: | 2020-01-27 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Mechanisms of activation and desensitization of full-length glycine receptor in lipid nanodiscs.
Nat Commun, 11, 2020
|
|
8QZD
 
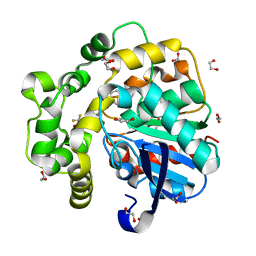 | | Soluble epoxide hydrolase in complex with Epoxykinin | | Descriptor: | 1,2-ETHANEDIOL, 2-[5-bromanyl-3-[2,2,2-tris(fluoranyl)ethanoyl]indol-1-yl]-N-cycloheptyl-ethanamide, BROMIDE ION, ... | | Authors: | Kumar, A, Ehrler, J.M.H, Ziegler, S, Doetsch, L, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-27 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of the sEH Inhibitor Epoxykynin as a Potent Kynurenine Pathway Modulator.
J.Med.Chem., 67, 2024
|
|
4V3J
 
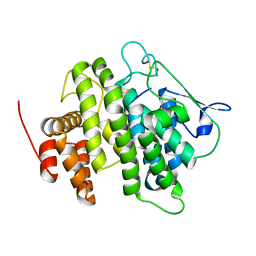 | | Structural and functional characterization of a novel monotreme- specific protein from the milk of the platypus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MONOTREME LACTATING PROTEIN | | Authors: | Kumar, A, Newman, J, Polekina, G, Adams, T.E, Sharp, J.A, Peat, T.S, Nicholas, K.R. | | Deposit date: | 2014-10-20 | | Release date: | 2016-01-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural characterization of a novel monotreme-specific protein with antimicrobial activity from the milk of the platypus.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6Y90
 
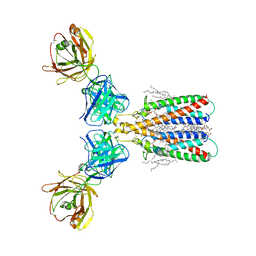 | |
6Y92
 
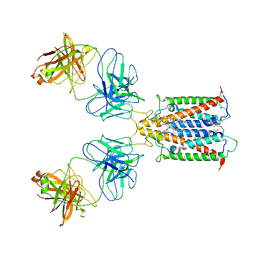 | |
6Y97
 
 | |
6Y9A
 
 | |
7NPW
 
 | |
5N2W
 
 | | WT-Parkin and pUB complex | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin,E3 ubiquitin-protein ligase parkin, Polyubiquitin-B, ... | | Authors: | Kumar, A, Chaugule, V.K, Johnson, C, Toth, R, Sundaramoorthy, R, Knebel, A, Walden, H. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5N38
 
 | | S65DParkin and pUB complex | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase parkin,E3 ubiquitin-protein ligase parkin, ... | | Authors: | Kumar, A, Chaugule, V.K, Johnson, C, Toth, R, Sundaramoorthy, R, Knebel, A, Walden, H. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6FC6
 
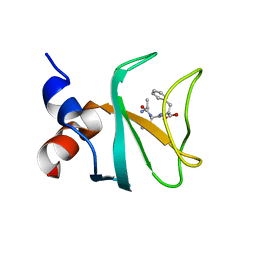 | |
5OAT
 
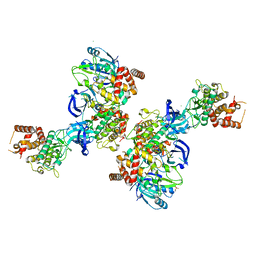 | | PINK1 structure | | Descriptor: | MAGNESIUM ION, Serine/threonine-protein kinase PINK1, mitochondrial-like Protein | | Authors: | Kumar, A, Tamjar, J, Woodroof, H.I, Raimi, O.G, Waddell, A.Y, Peggie, M, Muqit, M.M.K, van Aalten, D.M.F. | | Deposit date: | 2017-06-23 | | Release date: | 2017-10-11 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure of PINK1 and mechanisms of Parkinson's disease associated mutations.
Elife, 6, 2017
|
|
5N74
 
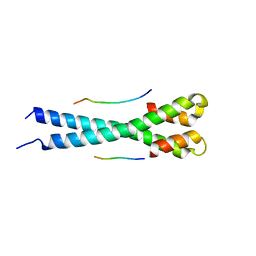 | | Microtubule end binding protein complex | | Descriptor: | Karyogamy protein KAR9, Microtubule-associated protein RP/EB family member 1 | | Authors: | Kumar, A, Steinmetz, M. | | Deposit date: | 2017-02-18 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Short Linear Sequence Motif LxxPTPh Targets Diverse Proteins to Growing Microtubule Ends.
Structure, 25, 2017
|
|
8C4A
 
 | |
1FMM
 
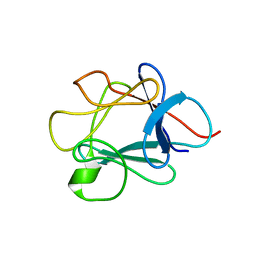 | | SOLUTION STRUCTURE OF NFGF-1 | | Descriptor: | ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | Arunkumar, A.I, Srisailam, S, Kumar, T.K.S, Chiu, I.M, Yu, C. | | Deposit date: | 2000-08-18 | | Release date: | 2001-08-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and stability of an acidic fibroblast growth factor from Notophthalmus viridescens.
J.Biol.Chem., 277, 2002
|
|
7Q4W
 
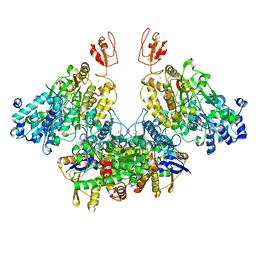 | | CryoEM structure of electron bifurcating Fe-Fe hydrogenase HydABC complex A. woodii in the oxidised state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Kumar, A, Saura, P, Poeverlein, M.C, Gamiz-Hernandez, A.P, Kaila, V.R.I, Mueller, V, Schuller, J.M. | | Deposit date: | 2021-11-02 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular Basis of the Electron Bifurcation Mechanism in the [FeFe]-Hydrogenase Complex HydABC.
J.Am.Chem.Soc., 145, 2023
|
|
8DK6
 
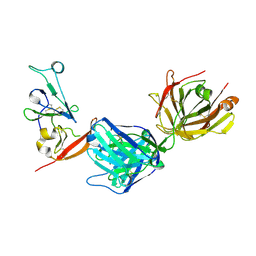 | | Structure of hepatitis C virus envelope N-terminal truncated glycoprotein 2 (E2) (residues 456-713) from J6 genotype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy chain, 2A12 Fab light chain, ... | | Authors: | Kumar, A, Rohe, T, Elrod, E.J, Khan, A.G, Dearborn, A.D, Kissinger, R, Grakoui, A, Marcotrigiano, J. | | Deposit date: | 2022-07-03 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Regions of hepatitis C virus E2 required for membrane association.
Nat Commun, 14, 2023
|
|
6VM3
 
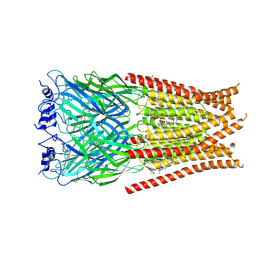 | | Full length Glycine receptor reconstituted in lipid nanodisc in Gly/IVM-conformation (State-3) | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kumar, A, Basak, S, Chakrapani, S. | | Deposit date: | 2020-01-27 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Mechanisms of activation and desensitization of full-length glycine receptor in lipid nanodiscs.
Nat Commun, 11, 2020
|
|
