3GDX
 
 | | Dna polymerase beta with a gapped DND substrate and dTMP(CF2)PP | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Wilson, S.H, Batra, V.K, Pedersen, L.C. | | Deposit date: | 2009-02-24 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Alpha,beta-difluoromethylene deoxynucleoside 5'-triphosphates: a convenient synthesis of useful probes for DNA polymerase beta structure and function
Org.Lett., 11, 2009
|
|
7UWM
 
 | | Structure of the IL-17A-IL-17RA binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor A, ... | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
7UWJ
 
 | | Structure of the homodimeric IL-25-IL-17RB binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor B, Interleukin-25 | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
7UWK
 
 | | Structure of the higher-order IL-25-IL-17RB complex | | Descriptor: | Interleukin-17 receptor B, Interleukin-25 | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
7UWN
 
 | | Structure of the IL-17A-IL-17RA-IL-17RC ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor A, ... | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
7QZK
 
 | |
7QZS
 
 | |
3G8O
 
 | | Progesterone Receptor with bound Pyrrolidine 1 | | Descriptor: | N~2~-[4-cyano-3-(trifluoromethyl)phenyl]-N,N-dimethyl-N~2~-(2,2,2-trifluoroethyl)-L-alaninamide, Progesterone receptor, SULFATE ION | | Authors: | Thompson, S.K, Washburn, D.G, Madauss, K.P, Williams, S.P, Stewart, E.L. | | Deposit date: | 2009-02-12 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational design of orally-active, pyrrolidine-based progesterone receptor partial agonists.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3BZK
 
 | |
3BEJ
 
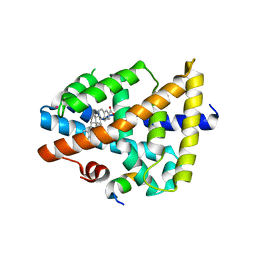 | | Structure of human FXR in complex with MFA-1 and co-activator peptide | | Descriptor: | (8alpha,10alpha,13alpha,17beta)-17-[(4-hydroxyphenyl)carbonyl]androsta-3,5-diene-3-carboxylic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Soisson, S.M, Parthasarathy, G, Becker, J.W. | | Deposit date: | 2007-11-19 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a potent synthetic FXR agonist with an unexpected mode of binding and activation.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4C5C
 
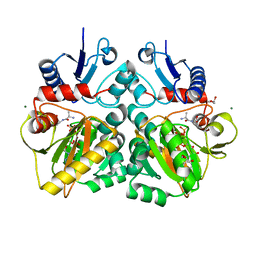 | | The X-ray crystal structure of D-alanyl-D-alanine ligase in complex with ADP and D-ala-D-ala | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-ALANINE, D-ALANINE--D-ALANINE LIGASE, ... | | Authors: | Batson, S, Majce, V, Lloyd, A.J, Rea, D, Fishwick, C.W.G, Simmons, K.J, Fulop, V, Roper, D.I. | | Deposit date: | 2013-09-10 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of D-Ala:D-Ala ligase through a phosphorylated form of the antibiotic D-cycloserine.
Nat Commun, 8, 2017
|
|
3Q7E
 
 | |
4C5A
 
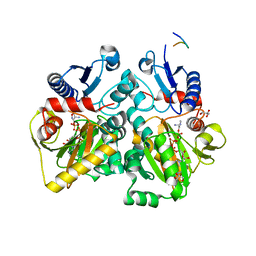 | | The X-ray crystal structures of D-alanyl-D-alanine ligase in complex ADP and D-cycloserine phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-ALANINE--D-ALANINE LIGASE, GLYCEROL, ... | | Authors: | Batson, S, Majce, V, Lloyd, A.J, Rea, D, Fishwick, C.W.G, Simmons, K.J, Fulop, V, Roper, D.I. | | Deposit date: | 2013-09-10 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibition of D-Ala:D-Ala ligase through a phosphorylated form of the antibiotic D-cycloserine.
Nat Commun, 8, 2017
|
|
3BZC
 
 | |
4C5B
 
 | | The X-ray crystal structure of D-alanyl-D-alanine ligase in complex with ATP and D-ala-D-ala | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBONATE ION, D-ALANINE, ... | | Authors: | Batson, S, Majce, V, Lloyd, A.J, Rea, D, Fishwick, C.W.G, Simmons, K.J, Fulop, V, Roper, D.I. | | Deposit date: | 2013-09-10 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibition of D-Ala:D-Ala ligase through a phosphorylated form of the antibiotic D-cycloserine.
Nat Commun, 8, 2017
|
|
2RJT
 
 | |
3R7O
 
 | | Structure of dually phosphorylated c-MET receptor kinase in complex with an MK-2461 analog | | Descriptor: | Hepatocyte growth factor receptor, N-[(2R)-1,4-dioxan-2-ylmethyl]-N-methyl-N'-{5-oxo-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl}sulfuric diamide | | Authors: | Soisson, S.M, Rickert, K, Patel, S.B, Munshi, S, Lumb, K.J. | | Deposit date: | 2011-03-22 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for selective small molecule kinase inhibition of activated c-Met.
J.Biol.Chem., 286, 2011
|
|
3ISS
 
 | | Crystal structure of enolpyruvyl-UDP-GlcNAc synthase (MurA):UDP-N-acetylmuramic acid:phosphite from Escherichia coli | | Descriptor: | PHOSPHITE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | Authors: | Jackson, S.G, Zhang, F, Chindemi, P, Junop, M.S, Berti, P.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evidence of Kinetic Control of Ligand Binding and Staged Product Release in MurA (Enolpyruvyl UDP-GlcNAc Synthase)-Catalyzed Reactions .
Biochemistry, 48, 2009
|
|
6EZM
 
 | | Imidazoleglycerol-phosphate dehydratase from Saccharomyces cerevisiae | | Descriptor: | Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION, [(2R)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl]phosphonic acid | | Authors: | Rawson, S, Bisson, C, Hurdiss, D.L, Muench, S.P. | | Deposit date: | 2017-11-15 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Elucidating the structural basis for differing enzyme inhibitor potency by cryo-EM.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EZJ
 
 | | Imidazoleglycerol-phosphate dehydratase | | Descriptor: | Imidazoleglycerol-phosphate dehydratase 2, chloroplastic, MANGANESE (II) ION, ... | | Authors: | Rawson, S, Bisson, C, Hurdiss, D.L, Muench, S.P. | | Deposit date: | 2017-11-15 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Elucidating the structural basis for differing enzyme inhibitor potency by cryo-EM.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3SCN
 
 | | Crystal Structure of Rice BGlu1 E386G Mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
3SCT
 
 | | Crystal Structure of Rice BGlu1 E386G Mutant Complexed with Cellotetraose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
3SCR
 
 | | Crystal Structure of Rice BGlu1 E386S Mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, GLYCEROL, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-06-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
3SCO
 
 | | Crystal Structure of Rice BGlu1 E386G Mutant Complexed with alpha-Glucosyl Fluoride | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, GLYCEROL, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
3SCV
 
 | | Crystal Structure of Rice BGlu1 E386G/S334A Mutant Complexed with Cellotetraose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
