8CAX
 
 | | Structure of Tau filaments Type II from Subacute Sclerosing Panencephalitis | | Descriptor: | Microtubule-associated protein tau | | Authors: | Qi, C, Hasegawa, M, Takao, M, Sakai, M, Akagi, M, Iwasaki, Y, Yoshida, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-01-24 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Identical tau filaments in subacute sclerosing panencephalitis and chronic traumatic encephalopathy.
Acta Neuropathol Commun, 11, 2023
|
|
8CAQ
 
 | | Structure of Tau filaments Type I from Subacute Sclerosing Panencephalitis | | Descriptor: | Microtubule-associated protein tau | | Authors: | Qi, C, Hasegawa, M, Takao, M, Sakai, M, Akagi, M, Iwasaki, Y, Yoshida, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-01-24 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Identical tau filaments in subacute sclerosing panencephalitis and chronic traumatic encephalopathy.
Acta Neuropathol Commun, 11, 2023
|
|
8BYN
 
 | | Chronic traumatic encephalopathy tau filaments with PET ligand flortaucipir | | Descriptor: | 7-(6-fluoranylpyridin-3-yl)-5~{H}-pyrido[4,3-b]indole, Microtubule-associated protein tau | | Authors: | Shi, Y, Ghetti, B, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2022-12-13 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM Structures of Chronic Traumatic Encephalopathy Tau Filaments with PET Ligand Flortaucipir.
J.Mol.Biol., 435, 2023
|
|
4V91
 
 | | Kluyveromyces lactis 80S ribosome in complex with CrPV-IRES | | Descriptor: | 25S RRNA, 5.8S RRNA, 5S RRNA, ... | | Authors: | Fernandez, I.S, Bai, X, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-03-21 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Initiation of Translation by Cricket Paralysis Virus Ires Requires its Translocation in the Ribosome.
Cell(Cambridge,Mass.), 157, 2014
|
|
6VPS
 
 | | Cryo-EM structure of the amyloid core of Drosophila Orb2 isolated from head | | Descriptor: | Translational regulator orb2 | | Authors: | Hervas, R, Rau, M.J, Park, Y, Zhang, W, Murzin, A.G, Fitzpatrick, J.A.J, Scheres, S.H.W, Si, K. | | Deposit date: | 2020-02-04 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM structure of a neuronal functional amyloid implicated in memory persistence in Drosophila
Science, 367, 2020
|
|
5A63
 
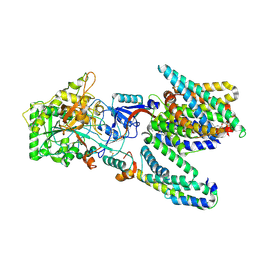 | | Cryo-EM structure of the human gamma-secretase complex at 3.4 angstrom resolution. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bai, X, Yan, C, Yang, G, Lu, P, Ma, D, Sun, L, Zhou, R, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An Atomic Structure of Human Gamma-Secretase
Nature, 525, 2015
|
|
9GG1
 
 | | P301T type I tau filaments from human brain | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Schweighauser, M, Shi, Y, Murzin, A.G, Garringer, H.J, Vidal, R, Murrell, J.R, Erro, M.E, Seelaar, H, Ferrer, I, van Swieten, J.C, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2024-08-12 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Novel tau filament folds in individuals with MAPT mutations P301L and P301T.
Biorxiv, 2024
|
|
9GG0
 
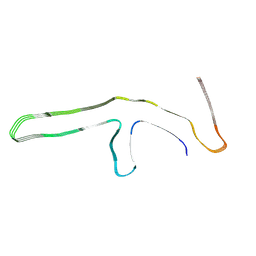 | | P301L tau filaments from human brain | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Schweighauser, M, Shi, Y, Murzin, A.G, Garringer, H.J, Vidal, R, Murrell, J.R, Erro, M.E, Seelaar, H, Ferrer, I, van Swieten, J.C, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2024-08-12 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Novel tau filament folds in individuals with MAPT mutations P301L and P301T.
Biorxiv, 2024
|
|
9GG6
 
 | | P301T type II tau filaments from human brain | | Descriptor: | Isoform Tau-F of Microtubule-associated protein tau | | Authors: | Schweighauser, M, Shi, Y, Murzin, A.G, Garringer, H.J, Vidal, R, Murrell, J.R, Erro, M.E, Seelaar, H, Ferrer, I, van Swieten, J.C, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2024-08-13 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Novel tau filament folds in individuals with MAPT mutations P301L and P301T.
Biorxiv, 2024
|
|
5FKW
 
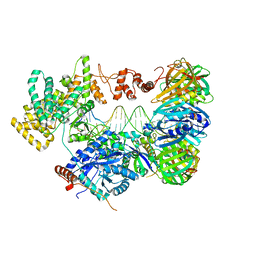 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon) | | Descriptor: | DNA POLYMERASE III ALPHA, DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
5FN5
 
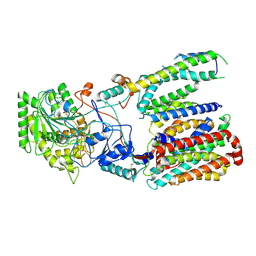 | | Cryo-EM structure of gamma secretase in class 3 of the apo- state ensemble | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2019-09-11 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
5FKV
 
 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon, tau complex) | | Descriptor: | DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, DNA POLYMERASE III SUBUNIT ALPHA, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.04 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
5M1S
 
 | | Cryo-EM structure of the E. coli replicative DNA polymerase-clamp-exonuclase-theta complex bound to DNA in the editing mode | | Descriptor: | DNA Primer Strand, DNA Template Strand, DNA polymerase III subunit alpha, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2016-10-10 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Self-correcting mismatches during high-fidelity DNA replication.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6OKK
 
 | |
6HRE
 
 | | Paired helical filament from sporadic Alzheimer's disease brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Falcon, B, Zhang, W, Schweighauser, M, Murzin, A.G, Vidal, R, Garringer, H.J, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2018-09-26 | | Release date: | 2018-10-10 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Tau filaments from multiple cases of sporadic and inherited Alzheimer's disease adopt a common fold.
Acta Neuropathol., 136, 2018
|
|
6HRF
 
 | | Straight filament from sporadic Alzheimer's disease brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Falcon, B, Zhang, W, Schweighauser, M, Murzin, A.G, Vidal, R, Garringer, H.J, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2018-09-26 | | Release date: | 2018-10-10 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Tau filaments from multiple cases of sporadic and inherited Alzheimer's disease adopt a common fold.
Acta Neuropathol., 136, 2018
|
|
6GX5
 
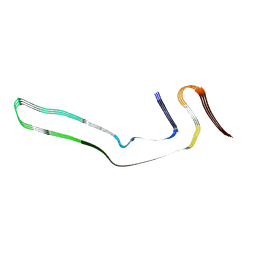 | | Narrow Pick Filament from Pick's disease brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Falcon, B, Zhang, W, Murzin, A.G, Murshudov, G, Garringer, H.J, Vidal, R, Crowther, R.A, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2018-06-26 | | Release date: | 2018-09-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of filaments from Pick's disease reveal a novel tau protein fold.
Nature, 561, 2018
|
|
4V92
 
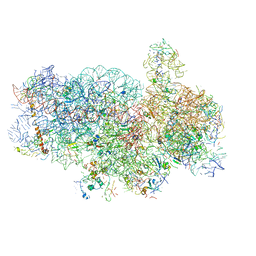 | | Kluyveromyces lactis 80S ribosome in complex with CrPV-IRES | | Descriptor: | 18S RRNA, ES1, ES10, ... | | Authors: | Fernandez, I.S, Bai, X, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-03-21 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Initiation of Translation by Cricket Paralysis Virus Ires Requires its Translocation in the Ribosome.
Cell(Cambridge,Mass.), 157, 2014
|
|
4V5X
 
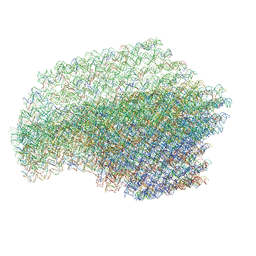 | | The cryo-EM structure of a 3D DNA-origami object | | Descriptor: | SCAFFOLD STRAND,SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Bai, X.C, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2012-10-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Cryo-Em Structure of a 3D DNA-Origami Object.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4V8Z
 
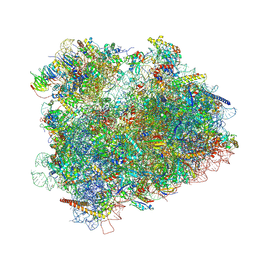 | | Cryo-EM reconstruction of the 80S-eIF5B-Met-itRNAMet Eukaryotic Translation Initiation Complex | | Descriptor: | 18S RIBOSOMAL RNA, 25S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN S0-A, ... | | Authors: | Fernandez, I.S, Bai, X.C, Hussain, T, Kelley, A.C, Lorsch, J.R, Ramakrishnan, V, Scheres, S.H.W. | | Deposit date: | 2013-07-20 | | Release date: | 2014-07-09 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Molecular architecture of a eukaryotic translational initiation complex.
Science, 342, 2013
|
|
4V8Y
 
 | | Cryo-EM reconstruction of the 80S-eIF5B-Met-itRNAMet Eukaryotic Translation Initiation Complex | | Descriptor: | 18S RIBOSOMAL RNA, 25S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN S0-A, ... | | Authors: | Fernandez, I.S, Bai, X.C, Hussain, T, Kelley, A.C, Lorsch, J.R, Ramakrishnan, V, Scheres, S.H.W. | | Deposit date: | 2013-07-20 | | Release date: | 2014-07-09 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular architecture of a eukaryotic translational initiation complex.
Science, 342, 2013
|
|
6XYO
 
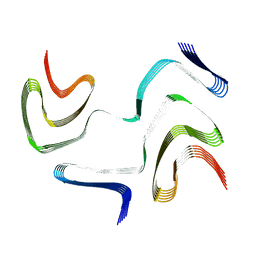 | | Multiple system atrophy Type I alpha-synuclein filament | | Descriptor: | Alpha-synuclein | | Authors: | Schweighauser, M, Shi, Y, Tarutani, A, Kametani, F, Murzin, A.G, Ghetti, B, Matsubara, T, Tomita, T, Ando, T, Hasegawa, K, Murayama, S, Yoshida, M, Hasegawa, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of alpha-synuclein filaments from multiple system atrophy.
Nature, 585, 2020
|
|
6XYP
 
 | | Multiple system atrophy Type II-1 alpha-synuclein filament | | Descriptor: | Alpha-synuclein | | Authors: | Schweighauser, M, Shi, Y, Tarutani, A, Kametani, F, Murzin, A.G, Ghetti, B, Matsubara, T, Tomita, T, Ando, T, Hasegawa, K, Murayama, S, Yoshida, M, Hasegawa, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structures of alpha-synuclein filaments from multiple system atrophy.
Nature, 585, 2020
|
|
6XYQ
 
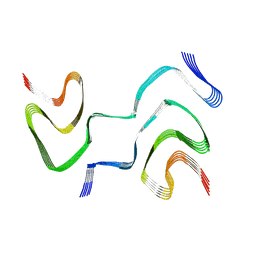 | | Multiple system atrophy Type II-2 alpha-synuclein filament | | Descriptor: | Alpha-synuclein | | Authors: | Schweighauser, M, Shi, Y, Tarutani, A, Kametani, F, Murzin, A.G, Ghetti, B, Matsubara, T, Tomita, T, Ando, T, Hasegawa, K, Murayama, S, Yoshida, M, Hasegawa, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structures of alpha-synuclein filaments from multiple system atrophy.
Nature, 585, 2020
|
|
5FKU
 
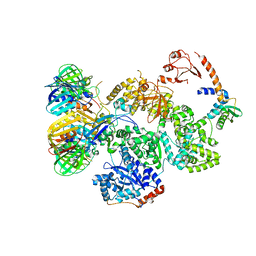 | | cryo-EM structure of the E. coli replicative DNA polymerase complex in DNA free state (DNA polymerase III alpha, beta, epsilon, tau complex) | | Descriptor: | DNA POLYMERASE III SUBUNIT ALPHA, DNA POLYMERASE III SUBUNIT BETA, DNA POLYMERASE III SUBUNIT EPSILON, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.34 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
