8G1A
 
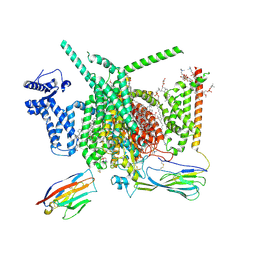 | | Cryo-EM structure of Nav1.7 with CBD | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cannabidiol inhibits Na v channels through two distinct binding sites.
Nat Commun, 14, 2023
|
|
8GTF
 
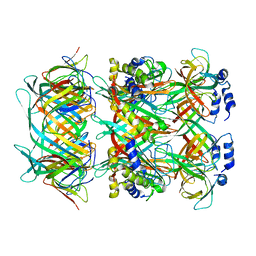 | | Cryo-EM model of the marine siphophage vB_DshS-R4C stopper-terminator complex | | Descriptor: | Head-to-tail joining protein, Major tail protein, Terminator protein | | Authors: | Huang, Y, Sun, H, Wei, S, Zheng, Q, Li, S, Zhang, R, Xia, N. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
7U3B
 
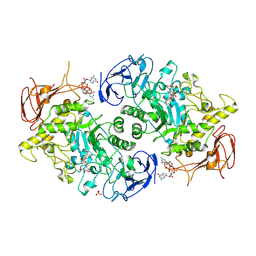 | | Structure of S. venezuelae GlgX bound to c-di-GMP and acarbose (pH 8.5) | | Descriptor: | 4-O-(4,6-dideoxy-4-{[(1S,2S,3S,4R,5S)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranosyl)-beta-D-glucopyranose, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Glycogen debranching enzyme GlgX, ... | | Authors: | Schumacher, M.A, Tschowri, N. | | Deposit date: | 2022-02-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Allosteric regulation of glycogen breakdown by the second messenger cyclic di-GMP.
Nat Commun, 13, 2022
|
|
8GCL
 
 | | Cryo-EM structure of hAQP2 in DDM | | Descriptor: | Aquaporin-2 | | Authors: | Kamegawa, A, Suzuki, S, Nishikawa, K, Numoto, N, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-03-02 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural analysis of the water channel AQP2 by single-particle cryo-EM.
J.Struct.Biol., 215, 2023
|
|
8WTM
 
 | |
5JPT
 
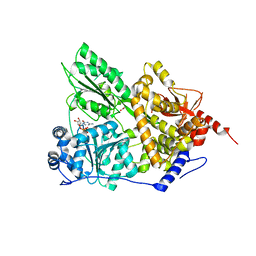 | | CRYSTAL STRUCTURE OF THE PRP43P DEAH-BOX RNA HELICASE IN COMPLEX WITH CDP | | Descriptor: | ACETATE ION, CYTIDINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Robert-Paganin, J, Rety, S, Leulliot, N. | | Deposit date: | 2016-05-04 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.935 Å) | | Cite: | Functional link between DEAH/RHA helicase Prp43 activation and ATP base binding.
Nucleic Acids Res., 45, 2017
|
|
8WTO
 
 | | Cryo-EM structure of jasmonic acid transporter ABCG16 in outward conformation | | Descriptor: | ABC transporter G family member 16, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Huang, X, An, N, Zhang, X, Zhang, P. | | Deposit date: | 2023-10-19 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM structure and molecular mechanism of the jasmonic acid transporter ABCG16
To Be Published
|
|
5KDR
 
 | | The crystal structure of carboxyltransferase from Staphylococcus Aureus bound to the antimicrobial agent moiramide B. | | Descriptor: | 1,2-ETHANEDIOL, Acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, Acetyl-coenzyme A carboxylase carboxyl transferase subunit beta, ... | | Authors: | Silvers, M.A, Pakhomova, S, Neau, D, Silvers, W.C, Anzalone, N, Taylor, C.M, Waldrop, G.L. | | Deposit date: | 2016-06-08 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Carboxyltransferase from Staphylococcus aureus Bound to the Antibacterial Agent Moiramide B.
Biochemistry, 55, 2016
|
|
5KEM
 
 | | EBOV sGP in complex with variable Fab domains of IgGs c13C6 and BDBV91 | | Descriptor: | BDBV91 variable Fab domain heavy chain, BDBV91 variable Fab domain light chain, Ebola secreted glycoprotein, ... | | Authors: | Pallesen, J, Murin, C.D, de Val, N, Cottrell, C.A, Hastie, K.M, Turner, H.L, Fusco, M.L, Flyak, A.I, Zeitlin, L, Crowe Jr, J.E, Andersen, K.G, Saphire, E.O, Ward, A.B. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structures of Ebola virus GP and sGP in complex with therapeutic antibodies.
Nat Microbiol, 1, 2016
|
|
5JXS
 
 | |
5JY6
 
 | |
8G01
 
 | | YES Complex - E. coli MraY, Protein E ID21, E. coli SlyD | | Descriptor: | FKBP-type peptidyl-prolyl cis-trans isomerase SlyD, GPE, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Orta, A.K, Clemons, W.M, Riera, N. | | Deposit date: | 2023-01-31 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The mechanism of the phage-encoded protein antibiotic from Phi X174.
Science, 381, 2023
|
|
8GL9
 
 | | Co-crystal structure of caPCNA bound to AOH1160 derivative 1LE | | Descriptor: | CHLORIDE ION, N~2~-(naphthalene-1-carbonyl)-N-(2-phenoxyphenyl)-L-alpha-glutamine, Proliferating cell nuclear antigen | | Authors: | Jossart, J, Kenjic, N, Malkas, L.H, Hickey, R.J, Perry, J.J. | | Deposit date: | 2023-03-21 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Small molecule targeting of transcription-replication conflict for selective chemotherapy.
Cell Chem Biol, 30, 2023
|
|
5K5C
 
 | | Structure of Mycobacterium thermoresistibile trehalose-6-phosphate synthase in a complex with Trehalose. | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Alpha,alpha-trehalose-phosphate synthase, ... | | Authors: | Mendes, V, Verma, N, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Mycobacterial OtsA Structures Unveil Substrate Preference Mechanism and Allosteric Regulation by 2-Oxoglutarate and 2-Phosphoglycerate.
Mbio, 10, 2019
|
|
8UUB
 
 | | Structure of hypothiocyanous acid reductase (Har) from Streptococcus pneumoniae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MALONATE ION, Oxidoreductase, ... | | Authors: | Shearer, H.L, Currie, M.J, Dickerhof, N, Dobson, R.C.J. | | Deposit date: | 2023-10-31 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hypothiocyanous acid reductase is critical for host colonization and infection by Streptococcus pneumoniae.
J.Biol.Chem., 300, 2024
|
|
5JUU
 
 | | Saccharomyces cerevisiae 80S ribosome bound with elongation factor eEF2-GDP-sordarin and Taura Syndrome Virus IRES, Structure V (least rotated 40S subunit) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Abeyrathne, P, Koh, C.S, Grant, T, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-05 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome.
Elife, 5, 2016
|
|
8GH4
 
 | | Complex of Adam 10 disentegrin cysteine rich domains with human monoclonal antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody heavy chain, Antibody light chain, ... | | Authors: | Nikolov, D.B, Saha, N, Xu, K, Goldgur, Y. | | Deposit date: | 2023-03-09 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Fully human monoclonal antibody targeting activated ADAM10 on colorectal cancer cells.
Biomed Pharmacother, 161, 2023
|
|
8G04
 
 | | Structure of signaling thrombopoietin-MPL receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Thrombopoietin, Thrombopoietin receptor, ... | | Authors: | Tsutsumi, N, Jude, K.M, Gati, C, Garcia, K.C. | | Deposit date: | 2023-01-31 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the thrombopoietin-MPL receptor complex is a blueprint for biasing hematopoiesis.
Cell, 186, 2023
|
|
7UVG
 
 | |
5LCM
 
 | | STRUCTURE OF the RAD14 DNA-binding domain IN COMPLEX WITH N2-acetylaminonaphtyl- GUANINE CONTAINING DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*CP*TP*AP*CP*(AAN)P*TP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*GP*TP*GP*AP*TP*GP*AP*CP*GP*TP*AP*GP*AP*G)-3'), DNA repair protein RAD14, ... | | Authors: | Schneider, S, Ebert, C, Simon, N, Carell, T. | | Deposit date: | 2016-06-22 | | Release date: | 2017-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into the Recognition of N(2) -Aryl- and C8-Aryl DNA Lesions by the Repair Protein XPA/Rad14.
Chembiochem, 18, 2017
|
|
8GQ1
 
 | | HyHEL10 Fab complexed with hen egg lysozyme carrying arginine cluster in framework region of light chain. | | Descriptor: | Heavy Chain of HyHel10 Antibody Fragment (Fab), Light Chain of HyHel10 Antibody Fragment (Fab), Lysozyme C, ... | | Authors: | Matsuura, H, Hirata, K, Sakai, N, Nakakido, M, Tsumoto, K. | | Deposit date: | 2022-08-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Arginine cluster introduction on framework region in anti-lysozyme antibody improved association rate constant by changing conformational diversity of CDR loops.
Protein Sci., 32, 2023
|
|
5LHE
 
 | |
7URP
 
 | |
7UY4
 
 | | Aminoglycoside-modifying enzyme ANT-3,9 in complex with spectinomycin and AMP-PNP | | Descriptor: | Aminoglycoside (3'') (9) adenylyltransferase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Reeve, S.M, Lee, R.E, Das, N, Jayaraman, S. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Characterization of the spectinomycin deactivating enzyme ANT-3,9
To Be Published
|
|
8GRT
 
 | | Small Dipeptide Analogues developed by Co-crystal Structure of Stenotrophomonas maltophilia Dipeptidyl Peptidase 7 | | Descriptor: | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID, Dipeptidyl-peptidase, TYROSINE | | Authors: | Yasumitsu, S, Koushi, H, Akihiro, N, Yoshiyuki, Y, Wataru, O, Mizuki, S, Saori, R, Nobutada, T, Anna, M, Keiko, H, Tsuda, Y. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Small Dipeptide Analogues Generated by Co-crystal Structure of Bacterial Dipeptidyl Peptidase 7 to Defeat Stenotrophomonas maltophilia
To Be Published
|
|
