6HU1
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 10 | | Descriptor: | 4-chloranyl-3-[(2,4-dichlorophenyl)carbonylamino]-~{N}-oxidanyl-benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
8A7Q
 
 | |
3MSL
 
 | |
7P60
 
 | |
7OKW
 
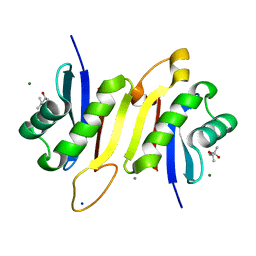 | | 1.62A X-ray crystal structure of the conserved C-terminal (CCT) of human OSR1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Bax, B.D, Elvers, K.T, Lipka-Lloyd, M, Mehellou, Y. | | Deposit date: | 2021-05-18 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structures of the Human SPAK and OSR1 Conserved C-Terminal (CCT) Domains.
Chembiochem, 23, 2022
|
|
4KXZ
 
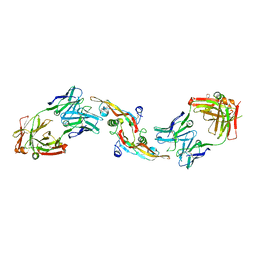 | | crystal structure of tgfb2 in complex with GC2008. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, GC1008 Heavy Chain, ... | | Authors: | Mathieu, M, Moulin, A.G, Wei, R. | | Deposit date: | 2013-05-28 | | Release date: | 2014-09-24 | | Last modified: | 2014-12-03 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structures of a pan-specific antagonist antibody complexed to different isoforms of TGF beta reveal structural plasticity of antibody-antigen interactions.
Protein Sci., 23, 2014
|
|
6HUV
 
 | | Yeast 20S proteasome with human beta2c (S171G) in complex with 39 | | Descriptor: | (2~{S})-~{N}-[(3~{S},4~{R})-1-cyclohexyl-5-methyl-4,5-bis(oxidanyl)hexan-3-yl]-3-(4-methoxyphenyl)-2-[[(2~{S})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-09 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
6W34
 
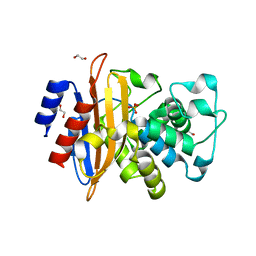 | | Crystal Structure of Class A Beta-lactamase from Bacillus cereus | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-08 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of Class A Beta-lactamase from
Bacillus cereus
To Be Published
|
|
4P1V
 
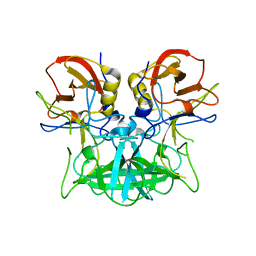 | | Structure of the P domain from a GI.7 Norovirus variant in complex with H-type 2 HBGA | | Descriptor: | P domain of VPI, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-02-27 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5497 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
6MU9
 
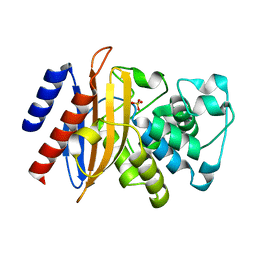 | |
1RL8
 
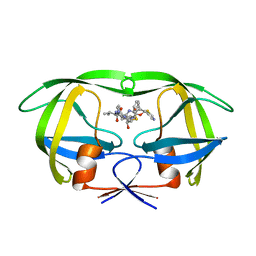 | | Crystal structure of the complex of resistant strain of hiv-1 protease(v82a mutant) with ritonavir | | Descriptor: | RITONAVIR, protease RETROPEPSIN | | Authors: | Rezacova, P, Brynda, J, Sedlacek, J, Konvalinka, J, Fabry, M, Horejsi, M. | | Deposit date: | 2003-11-25 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex of resistant strain of hiv-1 protease(v82a mutant) with ritonavir
To be Published, 2005
|
|
6W68
 
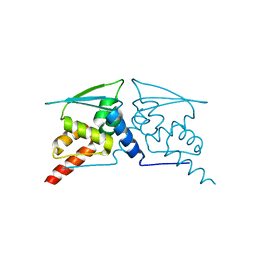 | |
6W18
 
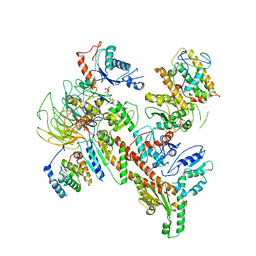 | | Structure of S. pombe Arp2/3 complex in inactive state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1, ... | | Authors: | Shaaban, M, Nolen, B.J, Chowdhury, S. | | Deposit date: | 2020-03-03 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM reveals the transition of Arp2/3 complex from inactive to nucleation-competent state.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W1U
 
 | | RT XFEL structure of Photosystem II 400 microseconds after the second illumination at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2NWQ
 
 | | Short chain dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | Probable short-chain dehydrogenase, SULFATE ION | | Authors: | McGrath, T.E, Kimber, M.S, Beattie, B, Thambipillai, D, Dharamsi, A, Virag, C, Nethery-Brokx, K, Edwards, A.M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2006-11-16 | | Release date: | 2006-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Short chain dehydrogenase from Pseudomonas aeruginosa
To be Published
|
|
6PWW
 
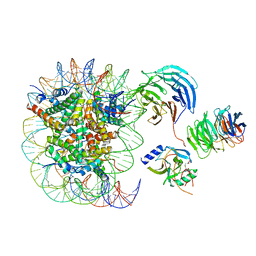 | | Cryo-EM structure of MLL1 in complex with RbBP5 and WDR5 bound to the nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
1RWH
 
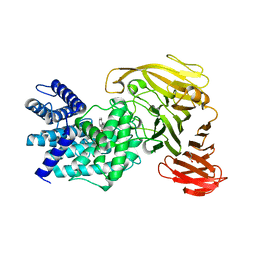 | | Crystal structure of Arthrobacter aurescens chondroitin AC lyase in complex with chondroitin tetrasaccharide | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-2,6-anhydro-3-deoxy-L-xylo-hexonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lunin, V.V, Li, Y, Miyazono, H, Kyogashima, M, Bell, A.W, Cygler, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
4PCD
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM ROSEOBACTER DENITRIFICANS OCh 114 (RD1_1052, TARGET EFI-510238) WITH BOUND L-GALACTONATE | | Descriptor: | C4-dicarboxylate transport system, substrate-binding protein, putative, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-15 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
6VZ1
 
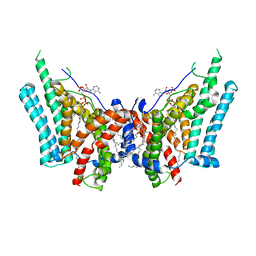 | | Cryo-EM structure of human diacylglycerol O-acyltransferase 1 complexed with acyl-CoA substrate | | Descriptor: | Diacylglycerol O-acyltransferase 1, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name), [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Sui, X, Wang, K, Gluchowski, N, Liao, M, Walther, C.T, Farese Jr, V.R. | | Deposit date: | 2020-02-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme.
Nature, 581, 2020
|
|
6VZ6
 
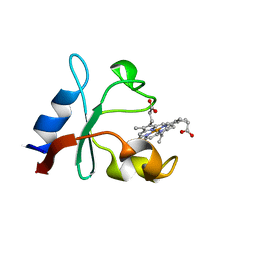 | | Methanococcoides burtonii cytochrome b5 domain protein (WP 011499504.1) | | Descriptor: | Cytochrome b5-domain protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Teakel, S.L, Forwood, J.K, Aragao, D, Cahill, M.A, Marama, M. | | Deposit date: | 2020-02-27 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methanococcoides burtonii cytochrome b5 domain protein (WP 011499504.1)
To Be Published
|
|
4PAK
 
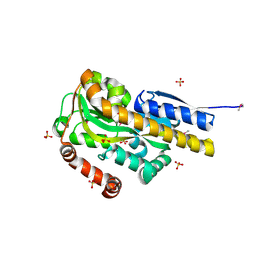 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM VERMINEPHROBACTER EISENIAE EF01-2 (Veis_3954, TARGET EFI-510324) A NEPHRIDIAL SYMBIONT OF THE EARTHWORM EISENIA FOETIDA, BOUND TO (R)-PANTOIC ACID | | Descriptor: | PANTOATE, SULFATE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-09 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
6VZ7
 
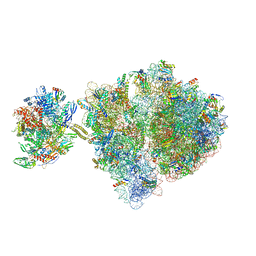 | | Escherichia coli transcription-translation complex C1 (TTC-C1) containing a 27 nt long mRNA spacer, NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
1UC4
 
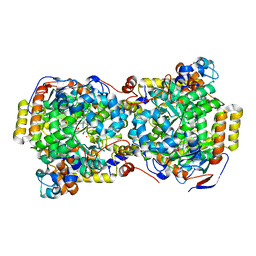 | | Structure of diol dehydratase complexed with (S)-1,2-propanediol | | Descriptor: | AMMONIUM ION, CYANOCOBALAMIN, POTASSIUM ION, ... | | Authors: | Shibata, N, Nakanishi, Y, Fukuoka, M, Yamanishi, M, Yasuoka, N, Toraya, T. | | Deposit date: | 2003-04-08 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural rationalization for the lack of stereospecificity in coenzyme B12-dependent diol dehydratase
J.BIOL.CHEM., 278, 2003
|
|
6JEB
 
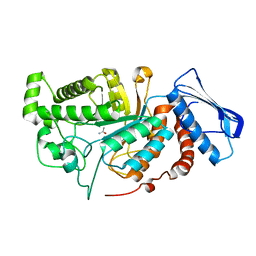 | | crystal structure of a beta-N-acetylhexosaminidase | | Descriptor: | ACETAMIDE, Beta-N-acetylhexosaminidase, ZINC ION | | Authors: | Chen, X, Wang, J.C, Liu, M.J, Yang, W.Y, Wang, Y.Z, Tang, R.P, Zhang, M. | | Deposit date: | 2019-02-05 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Crystallographic evidence for substrate-assisted catalysis of beta-N-acetylhexosaminidas from Akkermansia muciniphila.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
6W9C
 
 | | The crystal structure of papain-like protease of SARS CoV-2 | | Descriptor: | CHLORIDE ION, Non-structural protein 3, ZINC ION | | Authors: | Osipiuk, J, Jedrzejczak, R, Tesar, C, Endres, M, Stols, L, Babnigg, G, Kim, Y, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-22 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of papain-like protease of SARS CoV-2
to be published
|
|
