1MHW
 
 | | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides | | Descriptor: | 4-biphenylacetyl-Cys-(D)Arg-Tyr-N-(2-phenylethyl) amide, Cathepsin L | | Authors: | Chowdhury, S, Sivaraman, J, Wang, J, Devanathan, G, Lachance, P, Qi, H, Menard, R, Lefebvre, J, Konishi, Y, Cygler, M, Sulea, T, Purisima, E.O. | | Deposit date: | 2002-08-21 | | Release date: | 2002-12-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides
J.Med.Chem., 45, 2002
|
|
3BC3
 
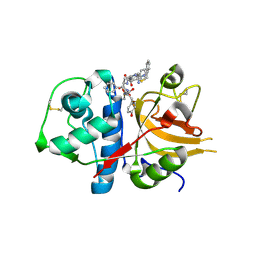 | | Exploring inhibitor binding at the S subsites of cathepsin L | | Descriptor: | Cathepsin L heavy and light chains, S-benzyl-N-(biphenyl-4-ylacetyl)-L-cysteinyl-N~5~-(diaminomethyl)-D-ornithyl-N-(2-phenylethyl)-L-tyrosinamide | | Authors: | Chowdhury, S.F, Joseph, L, Kumar, S, Tulsidas, S.R, Bhat, S, Ziomek, E, Nard, R.M, Sivaraman, J, Purisima, E.O. | | Deposit date: | 2007-11-12 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring inhibitor binding at the S' subsites of cathepsin L
J.Med.Chem., 51, 2008
|
|
6NE0
 
 | | Structure of double-stranded target DNA engaged Csy complex from Pseudomonas aeruginosa (PA-14) | | Descriptor: | CRISPR RNA (60-MER), CRISPR target DNA (44-MER), CRISPR-associated endonuclease Cas6/Csy4, ... | | Authors: | Chowdhury, S, Rollins, M.F, Carter, J, Golden, S.M, Miettinen, H.M, Santiago-Frangos, A, Faith, D, Lawrence, M.C, Wiedenheft, B, Lander, G.C. | | Deposit date: | 2018-12-15 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals a Mechanism of CRISPR-RNA-Guided Nuclease Recruitment and Anti-CRISPR Viral Mimicry.
Mol. Cell, 74, 2019
|
|
5JZM
 
 | |
5UZ9
 
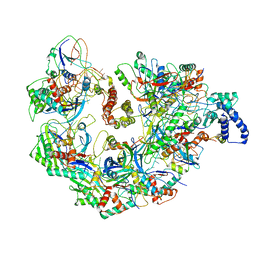 | | Cryo EM structure of anti-CRISPRs, AcrF1 and AcrF2, bound to type I-F crRNA-guided CRISPR surveillance complex | | Descriptor: | Anti-CRISPR protein 30, Anti-CRISPR protein Acr30-35, CRISPR RNA (60-MER), ... | | Authors: | Chowdhury, S, Carter, J, Rollins, M.F, Jackson, R.N, Hoffmann, C, Nosaka, L, Bondy-Denomy, J, Maxwell, K.L, Davidson, A.R, Fischer, E.R, Lander, G.C, Wiedenheft, B. | | Deposit date: | 2017-02-25 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals Mechanisms of Viral Suppressors that Intercept a CRISPR RNA-Guided Surveillance Complex.
Cell, 169, 2017
|
|
2GIO
 
 | |
2GIP
 
 | |
5JZO
 
 | |
5K4A
 
 | |
4WK5
 
 | | Crystal structure of a Isoprenoid Synthase family member from Thermotoga neapolitana DSM 4359, target EFI-509458 | | Descriptor: | Geranyltranstransferase | | Authors: | Toro, R, Bhosle, R, Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Poulter, C.D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a Isoprenoid Synthase family member from Thermotoga neapolitana DSM 4359, target EFI-509458
To be published
|
|
4WUT
 
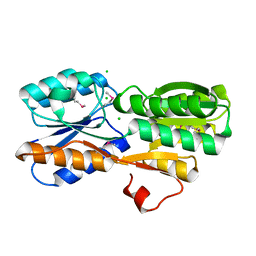 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS (Avi_5133, TARGET EFI-511220) WITH BOUND D-FUCOSE | | Descriptor: | ABC transporter substrate binding protein (Ribose), CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-03 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS (Avi_5133, TARGET EFI-511220) WITH BOUND D-FUCOSE
To be published
|
|
8FF2
 
 | |
6W17
 
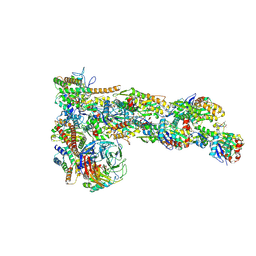 | | Structure of Dip1-activated Arp2/3 complex with nucleated actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Shaaban, M, Nolen, B.J, Chowdhury, S. | | Deposit date: | 2020-03-03 | | Release date: | 2020-08-12 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM reveals the transition of Arp2/3 complex from inactive to nucleation-competent state.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W18
 
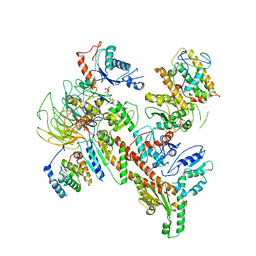 | | Structure of S. pombe Arp2/3 complex in inactive state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1, ... | | Authors: | Shaaban, M, Nolen, B.J, Chowdhury, S. | | Deposit date: | 2020-03-03 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM reveals the transition of Arp2/3 complex from inactive to nucleation-competent state.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3M0E
 
 | | Crystal structure of the ATP-bound state of Walker B mutant of NtrC1 ATPase domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Transcriptional regulator (NtrC family) | | Authors: | Chen, B, Sysoeva, T.A, Chowdhury, S, Rusu, M, Birmanns, S, Guo, L, Hanson, J, Yang, H, Nixon, B.T. | | Deposit date: | 2010-03-02 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Engagement of Arginine Finger to ATP Triggers Large Conformational Changes in NtrC1 AAA+ ATPase for Remodeling Bacterial RNA Polymerase.
Structure, 18, 2010
|
|
5HFK
 
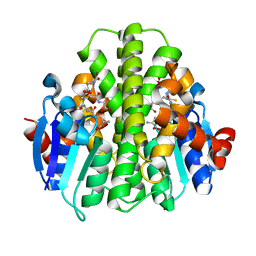 | | CRYSTAL STRUCTURE OF A GLUTATHIONE S-TRANSFERASE PROTEIN FROM ESCHERICHIA COLI OCh 157:H7 STR. SAKAI (ECs3186, TARGET EFI-507414) WITH BOUND GLUTATHIONE | | Descriptor: | Disulfide-bond oxidoreductase YfcG, GLUTATHIONE | | Authors: | Himmel, D.M, Toro, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | CRYSTAL STRUCTURE OF A GLUTATHIONE S-TRANSFERASE PROTEIN FROM ESCHERICHIA COLI OCh 157:H7 STR. SAKAI (ECs3186, TARGET EFI-507414) WITH BOUND GLUTATHIONE
TO BE PUBLISHED
|
|
7EZJ
 
 | | Crystal structure of p73 DNA binding domain complex bound with 1 bp and 2 bp spacer DNA response elements. | | Descriptor: | 12-mer DNA, Tumor protein p73, ZINC ION | | Authors: | Koley, T, Roy Chowdhury, S, Kumar, M, Kaur, P, Singh, T.P, Viadiu, H, Ethayathulla, A.S. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Deciphering the mechanism of p73 recognition of p53 response elements using the crystal structure of p73-DNA complexes and computational studies.
Int.J.Biol.Macromol., 206, 2022
|
|
3H8C
 
 | | A combined crystallographic and molecular dynamics study of cathepsin-L retro-binding inhibitors (compound 14) | | Descriptor: | Cathepsin L1, N-(biphenyl-4-ylacetyl)-S-methyl-L-cysteinyl-D-arginyl-N-(2-phenylethyl)-L-phenylalaninamide | | Authors: | Tulsidas, S.R, Chowdhury, S.F, Kumar, S, Joseph, L, Purisima, E.O, Sivaraman, J. | | Deposit date: | 2009-04-29 | | Release date: | 2009-10-20 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Combined Crystallographic and Molecular Dynamics Study of Cathepsin L Retrobinding Inhibitors
J.Med.Chem., 2009
|
|
7USC
 
 | | Cryo-EM structure of WAVE Regulatory Complex | | Descriptor: | Abl interactor 2, Cytoplasmic FMR1-interacting protein 1, Nck-associated protein 1, ... | | Authors: | Ding, B, Yang, S, Chen, B, Chowdhury, S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-09-21 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures reveal a key mechanism of WAVE regulatory complex activation by Rac1 GTPase.
Nat Commun, 13, 2022
|
|
7USD
 
 | | Cryo-EM structure of D-site Rac1-bound WAVE Regulatory Complex | | Descriptor: | Abl interactor 2, Cytoplasmic FMR1-interacting protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ding, B, Yang, S, Chen, B, Chowdhury, S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-09-21 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures reveal a key mechanism of WAVE regulatory complex activation by Rac1 GTPase.
Nat Commun, 13, 2022
|
|
7USE
 
 | | Cryo-EM structure of WAVE regulatory complex with Rac1 bound on both A and D site | | Descriptor: | Abl interactor 2, Cytoplasmic FMR1-interacting protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ding, B, Yang, S, Chen, B, Chowdhury, S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-09-21 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures reveal a key mechanism of WAVE regulatory complex activation by Rac1 GTPase.
Nat Commun, 13, 2022
|
|
7TPT
 
 | | Single-particle Cryo-EM structure of Arp2/3 complex at branched-actin junction. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Ding, B, Narvaez-Ortiz, H.Y, Nolen, B.J, Chowdhury, S. | | Deposit date: | 2022-01-26 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of Arp2/3 complex at a branched actin filament junction resolved by single-particle cryo-electron microscopy.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3H8B
 
 | | A combined crystallographic and molecular dynamics study of cathepsin-L retro-binding inhibitors(compound 9) | | Descriptor: | Cathepsin L1, N~2~,N~6~-bis(biphenyl-4-ylacetyl)-L-lysyl-D-arginyl-N-(2-phenylethyl)-L-phenylalaninamide | | Authors: | Tulsidas, S.R, Chowdhury, S.F, Kumar, S, Joseph, L, Purisima, E.O, Sivaraman, J. | | Deposit date: | 2009-04-29 | | Release date: | 2009-10-20 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A combined crystallographic and molecular dynamics study of cathepsin L retrobinding inhibitors
J.Med.Chem., 52, 2009
|
|
3H89
 
 | | A combined crystallographic and molecular dynamics study of cathepsin-L retro-binding inhibitors(compound 4) | | Descriptor: | Cathepsin L1, N~2~,N~6~-bis(biphenyl-4-ylacetyl)-L-lysyl-D-arginyl-N-(2-phenylethyl)-L-tyrosinamide | | Authors: | Tulsidas, S.R, Chowdhury, S.F, Kumar, S, Joseph, L, Purisima, E.O, Sivaraman, J. | | Deposit date: | 2009-04-29 | | Release date: | 2009-10-20 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A combined crystallographic and molecular dynamics study of cathepsin L retrobinding inhibitors
J.Med.Chem., 52, 2009
|
|
7KX7
 
 | | Cryo-EM structure of Ephydatia fluviatilis PiwiA-piRNA complex | | Descriptor: | MAGNESIUM ION, Piwi-A, RNA (5'-R(P*UP*CP*UP*CP*AP*GP*(OMC))-3') | | Authors: | Anzelon, T.A, Chowdhury, S, Lander, G.C, MacRae, I.J. | | Deposit date: | 2020-12-03 | | Release date: | 2021-07-14 | | Last modified: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for piRNA targeting.
Nature, 597, 2021
|
|
