4O60
 
 | | Structure of ankyrin repeat protein | | Descriptor: | ANK-N5C-317 | | Authors: | Ethayathulla, A.S, Guan, L. | | Deposit date: | 2013-12-20 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | A transcription blocker isolated from a designed repeat protein combinatorial library by in vivo functional screen.
Sci Rep, 5, 2015
|
|
3VD0
 
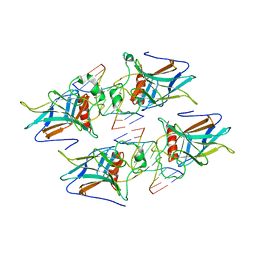 | | structure of p73 DNA binding domain tetramer modulates p73 transactivation | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*CP*AP*TP*GP*CP*CP*TP*G)-3'), Tumor protein p73, ZINC ION | | Authors: | Ethayathulla, A.S, Tse, P.W, Nguyen, S, Viadiu, H. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of p73 DNA-binding domain tetramer modulates p73 transactivation.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VD1
 
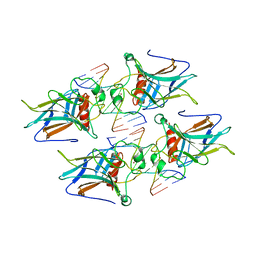 | | structure of p73 DNA binding domain tetramer modulates p73 transactivation | | Descriptor: | DNA (5'-D(*CP*GP*GP*GP*CP*AP*TP*GP*CP*CP*CP*G)-3'), Tumor protein p73, ZINC ION | | Authors: | Ethayathulla, A.S, Tse, P.W, Nguyen, S, Viadiu, H. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of p73 DNA-binding domain tetramer modulates p73 transactivation.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VD2
 
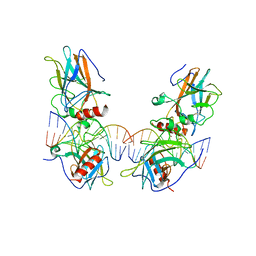 | | structure of p73 DNA binding domain tetramer modulates p73 transactivation | | Descriptor: | DNA (5'-D(*AP*TP*GP*GP*AP*CP*AP*TP*GP*TP*CP*CP*AP*T)-3'), Tumor protein p73, ZINC ION | | Authors: | Ethayathulla, A.S, Tse, P.W, Nguyen, S, Viadiu, H. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of p73 DNA-binding domain tetramer modulates p73 transactivation.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4G82
 
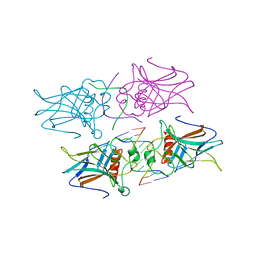 | |
4G83
 
 | |
4GUQ
 
 | |
2G8Z
 
 | | Crystal structure of the ternary complex of signalling protein from sheep (SPS-40) with trimer and designed peptide at 2.5A resolution | | Descriptor: | (TRP)(PRO)(TRP), 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, ... | | Authors: | Ethayathulla, A.S, Srivastava, D.B, Kumar, J, Somvanshi, R.K, Sharma, S, Singh, T.P. | | Deposit date: | 2006-03-04 | | Release date: | 2006-04-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the ternary complex of signalling protein from sheep (SPS-40) with trimer and designed peptide at 2.5A resolution
To be Published
|
|
1ZU8
 
 | | Crystal structure of the goat signalling protein with a bound trisaccharide reveals that Trp78 reduces the carbohydrate binding site to half | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3 like protein 1, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ethayathulla, A.S, Kumar, J, Srivastava, D.B, Singh, N, Sharma, S, Bhushan, A, Singh, T.P. | | Deposit date: | 2005-05-30 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of the goat signalling protein with a bound trisaccharide reveals that Trp78 reduces the carbohydrate binding site to half
To be Published
|
|
1ZL1
 
 | | Crystal structure of the complex of signalling protein from sheep (SPS-40) with a designed peptide Trp-His-Trp reveals significance of Asn79 and Trp191 in the complex formation | | Descriptor: | Chitinase-3 like protein 1, TRP-HIS-TRP peptide, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ethayathulla, A.S, Srivastava, D.B, Singh, N, Kumar, J, Somvanshi, R.K, Sharma, S, Dey, S, Singh, T.P. | | Deposit date: | 2005-05-05 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the complex of signalling protein from sheep (SPS-40) with a designed peptide Trp-His-Trp reveals significance of Asn79 and Trp191 in the complex formation
TO BE PUBLISHED
|
|
1SKG
 
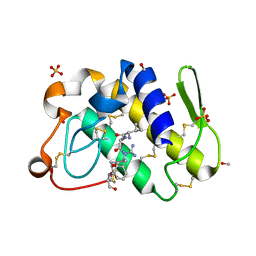 | | Structure-based rational drug design: Crystal structure of the complex formed between Phospholipase A2 and a pentapeptide Val-Ala-Phe-Arg-Ser | | Descriptor: | METHANOL, Phospholipase A2, SULFATE ION, ... | | Authors: | Ethayathulla, A.S, Singh, N, Sharma, S, Makker, J, Dey, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2004-03-04 | | Release date: | 2004-04-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structure-based rational drug design: Crystal structure of the complex formed between Phospholipase A2 and a pentapeptide Val-Ala-Phe-Arg-Ser
To be Published
|
|
4GUO
 
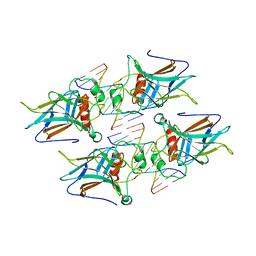 | |
4M64
 
 | |
2DT2
 
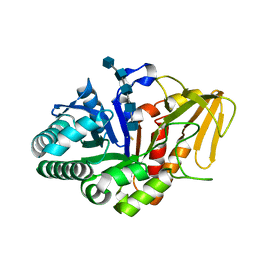 | | Crystal structure of the complex formed between goat signalling protein with pentasaccharide at 2.9A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1 | | Authors: | Ethayathulla, A.S, Kumar, J, Srivastava, D.B, Singh, N, Sharma, S, Bhushan, A, Singh, T.P. | | Deposit date: | 2006-07-09 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Carbohydrate-binding properties of goat secretory glycoprotein (SPG-40) and its functional implications: structures of the native glycoprotein and its four complexes with chitin-like oligosaccharides
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2A7T
 
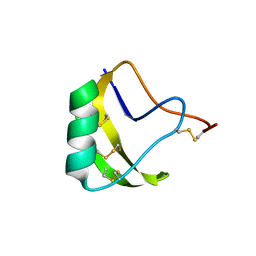 | | Crystal Structure of a novel neurotoxin from Buthus tamalus at 2.2A resolution. | | Descriptor: | Neurotoxin | | Authors: | Ethayathulla, A.S, Sharma, M, Saravanan, K, Sharma, S, Kaur, P, Yadav, S, Srinivasan, A, Singh, T.P. | | Deposit date: | 2005-07-06 | | Release date: | 2005-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a highly acidic neurotoxin from scorpion Buthus tamulus at 2.2A resolution reveals novel structural features.
J.Struct.Biol., 155, 2006
|
|
2B31
 
 | | Crystal structure of the complex formed between goat signalling protein with pentasaccharide at 3.1 A resolution reveals large scale conformational changes in the residues of TIM barrel | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, ... | | Authors: | Ethayathulla, A.S, Kumar, J, Srivastava, D.B, Singh, N, Sharma, S, Bhushan, A, Singh, T.P. | | Deposit date: | 2005-09-19 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the complex formed between goat signalling protein with pentasaccharide at 3.1 A resolution reveals large scale conformational changes in the residues of TIM barrel
TO BE PUBLISHED
|
|
4QFV
 
 | |
2O9O
 
 | | Crystal Structure of the buffalo Secretory Signalling Glycoprotein at 2.8 A resolution | | Descriptor: | Chitinase-3-like protein 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ethayathulla, A.S, Srivastava, D.B, Kumar, J, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2006-12-14 | | Release date: | 2007-01-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the buffalo secretory signalling glycoprotein at 2.8 A resolution
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2PQ2
 
 | | Structure of serine proteinase K complex with a highly flexible hydrophobic peptide at 1.8A resolution | | Descriptor: | CALCIUM ION, GALAG peptide, NITRATE ION, ... | | Authors: | Ethayathulla, A.S, Singh, A.K, Singh, N, Sharma, S, Sinha, M, Somvanshi, R.K, Kaur, P, Dey, S, Srinivasan, A, Singh, T.P. | | Deposit date: | 2007-05-01 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of serine proteinase K complex with a highly flexible hydrophobic peptide at 1.8A resolution
To be Published
|
|
2YZ2
 
 | | Crystal structure of the ABC transporter in the cobalt transport system | | Descriptor: | Putative ABC transporter ATP-binding protein TM_0222 | | Authors: | Ethayathullah, A.S, Bessho, Y, Padmanabhan, B, Singh, T.P, Kaur, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-02 | | Release date: | 2007-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the ABC transporter in the cobalt transport system
To be Published
|
|
2YYZ
 
 | | Crystal structure of Sugar ABC transporter, ATP-binding protein | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, Sugar ABC transporter, ... | | Authors: | Ethayathullah, A.S, Bessho, Y, Padmanabhan, B, Singh, T.P, Kaur, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-02 | | Release date: | 2007-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of Sugar ABC transporter, ATP-binding protein
To be Published
|
|
2ESC
 
 | | Crystal structure of a 40 KDa protective signalling protein from Bovine (SPC-40) at 2.1 A resolution | | Descriptor: | Chitinase-3-like protein 1, alpha-D-mannopyranose-(1-4)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kumar, J, Ethayathulla, A.S, Srivastav, D.B, Sharma, S, Srinivasan, A, Singh, T.P. | | Deposit date: | 2005-10-26 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a bovine secretory signalling glycoprotein (SPC-40) at 2.1 Angstrom resolution.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
8HCJ
 
 | | Structure of GH43 family enzyme, Xylan 1, 4 Beta- xylosidase from pseudopedobacter saltans | | Descriptor: | CALCIUM ION, Xylan 1,4-beta-xylosidase | | Authors: | Vishwakarma, P, Sachdeva, E, Goyal, A, Ethayathulla, A.S, Das, U, Kaur, P. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.566 Å) | | Cite: | Structure of GH43 family enzyme, Xylan 1, 4 Beta- xylosidase from pseudopedobacter saltans
To Be Published
|
|
7EZJ
 
 | | Crystal structure of p73 DNA binding domain complex bound with 1 bp and 2 bp spacer DNA response elements. | | Descriptor: | 12-mer DNA, Tumor protein p73, ZINC ION | | Authors: | Koley, T, Roy Chowdhury, S, Kumar, M, Kaur, P, Singh, T.P, Viadiu, H, Ethayathulla, A.S. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Deciphering the mechanism of p73 recognition of p53 response elements using the crystal structure of p73-DNA complexes and computational studies.
Int.J.Biol.Macromol., 206, 2022
|
|
6J90
 
 | | Crystal Structure of GyraseB N-Terminal Domain complex with ATP from Salmonella Typhi at 2.2A Resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kaur, G, Sachdeva, E, Tiwari, P, Gupta, D, Ethayathulla, A.S, Kaur, P. | | Deposit date: | 2019-01-21 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of GyraseB N-Terminal Domain complex with ATP from Salmonella Typhi at 2.2A Resolution
To Be Published
|
|
