5J3H
 
 | | Human insulin receptor domains L1-CR in complex with peptide S519C16 and 83-7 Fv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lawrence, M, Menting, J, Lawrence, C. | | Deposit date: | 2016-03-30 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Insulin Mimetic Peptide Disrupts the Primary Binding Site of the Insulin Receptor.
J.Biol.Chem., 291, 2016
|
|
3KIC
 
 | |
4V1M
 
 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | Descriptor: | 5'-D(*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP)-3', 5'-D(*CP*CP*AP*GP*GP*AP)-3', 5'-D(*TP*CP*AP*AP*GP*TP*AP*CP*TP*TP*TP*TP*TP*CP *CP*BRUP*GP*GP*TP*C)-3', ... | | Authors: | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
5IUX
 
 | | GLIC-V135C bimane labelled X-ray structure | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2,3,5,6-tetramethyl-1H,7H-pyrazolo[1,2-a]pyrazole-1,7-dione, ACETATE ION, ... | | Authors: | Fourati, Z, Menny, A, Delarue, M. | | Deposit date: | 2016-03-18 | | Release date: | 2017-03-29 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a pre-active conformation of a pentameric channel receptor.
Elife, 6, 2017
|
|
3KMT
 
 | | Crystal structure of vSET/SAH/H3 ternary complex | | Descriptor: | A612L protein, Histone H3, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wei, H, Zhou, M.M. | | Deposit date: | 2009-11-11 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dimerization of a viral SET protein endows its function.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6ZH0
 
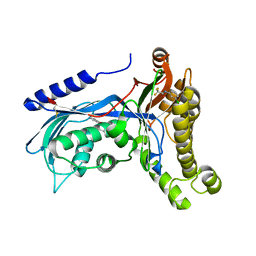 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, N-(3-chlorophenyl)-2,2,2-trifluoroacetamide, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
4V5N
 
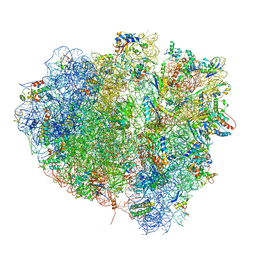 | | tRNA translocation on the 70S ribosome: the post- translocational translocation intermediate TI(POST) | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ratje, A.H, Loerke, J, Mikolajka, A, Bruenner, M, Hildebrand, P.W, Starosta, A.L, Doenhoefer, A, Connell, S.R, Fucini, P, Mielke, T, Whitford, P.C, Onuchic, J.N, Yu, Y, Sanbonmatsu, K.Y, Hartmann, R.K, Penczek, P.A, Wilson, D.N, Spahn, C.M.T. | | Deposit date: | 2010-10-21 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Head Swivel on the Ribosome Facilitates Translocation by Means of Intra-Subunit tRNA Hybrid Sites.
Nature, 468, 2010
|
|
3KIE
 
 | |
3KLW
 
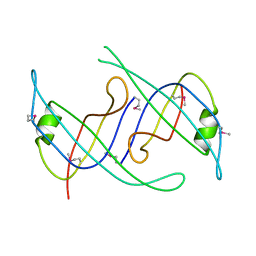 | | Crystal structure of primosomal replication protein n from Bordetella pertussis. northeast structural genomics consortium target ber132. | | Descriptor: | Primosomal replication protein n | | Authors: | Kuzin, A.P, Neely, H, Vorobiev, S.M, Seetharaman, J, Forouhar, F, Wang, H, Janjua, H, Maglaqui, M, Xiao, T, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-11-09 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of primosomal replication protein n from Bordetella pertussis. northeast structural genomics consortium target ber132.
To be Published
|
|
6ZMO
 
 | | SARS-CoV-2 Nsp1 bound to the human LYAR-80S-eEF1a ribosome complex | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
3KPQ
 
 | | Crystal Structure of HLA B*4405 in complex with EEYLKAWTF, a mimotope | | Descriptor: | Beta-2-microglobulin, EEYLKAWTF, mimotope peptide, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
6ZMI
 
 | | SARS-CoV-2 Nsp1 bound to the human LYAR-80S ribosome complex | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-02 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
3KKM
 
 | | Crystal structure of H-Ras T35S in complex with GppNHp | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Muraoka, S, Shima, F, Liao, J, Ijiri, Y, Matsumoto, K, Ye, M, Inoue, T, Kataoka, T. | | Deposit date: | 2009-11-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for conformational dynamics of GTP-bound Ras protein
J.Biol.Chem., 285, 2010
|
|
3KWJ
 
 | | Structure of human DPP-IV with (2S,3S,11bS)-3-(3-Fluoromethyl-phenyl)-9,10-dimethoxy-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-2-ylamine | | Descriptor: | (2S,3S,11bS)-3-[3-(fluoromethyl)phenyl]-9,10-dimethoxy-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 soluble form | | Authors: | Hennig, M, Stihle, M, Thoma, R. | | Deposit date: | 2009-12-01 | | Release date: | 2010-09-15 | | Last modified: | 2020-09-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Aryl- and heteroaryl-substituted aminobenzo[a]quinolizines as dipeptidyl peptidase IV inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6ZM7
 
 | | SARS-CoV-2 Nsp1 bound to the human CCDC124-80S-EBP1 ribosome complex | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-01 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
1QJY
 
 | | HUMAN RHINOVIRUS 16 COAT PROTEIN IN COMPLEX WITH ANTIVIRAL COMPOUND VP65099 | | Descriptor: | 2,6-DIMETHYL-1-(3-[3-METHYL-5-ISOXAZOLYL]-PROPANYL)-4-[2-METHYL-4-ISOXAZOLYL]-PHENOL, MYRISTIC ACID, PROTEIN VP1, ... | | Authors: | Hadfield, A.T, Diana, G.D, Rossmann, M.G. | | Deposit date: | 1999-07-06 | | Release date: | 1999-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of Three Structurally Related Antiviral Compounds in Complex with Human Rhinovirus 16
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1HPO
 
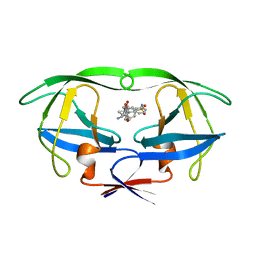 | | HIV-1 PROTEASE TRIPLE MUTANT/U103265 COMPLEX | | Descriptor: | 4-CYANO-N-(3-CYCLOPROPYL(5,6,7,8,9,10-HEXAHYDRO-4-HYDROXY-2-OXO-CYCLOOCTA[B]PYRAN-3-YL)METHYL)PHENYL BENZENSULFONAMIDE, HIV-1 PROTEASE | | Authors: | Watenpaugh, K.D, Janakiraman, M.N. | | Deposit date: | 1996-12-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based design of nonpeptidic HIV protease inhibitors: the sulfonamide-substituted cyclooctylpyramones.
J.Med.Chem., 40, 1997
|
|
5J7C
 
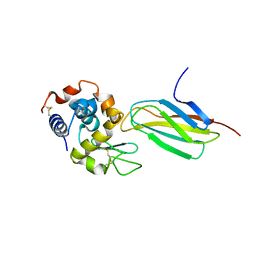 | |
6ZG1
 
 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
1GPD
 
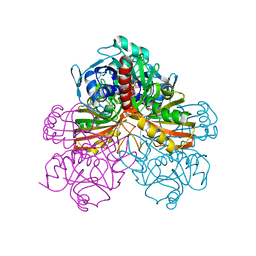 | | STUDIES OF ASYMMETRY IN THE THREE-DIMENSIONAL STRUCTURE OF LOBSTER D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE | | Descriptor: | D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Moras, D, Olsen, K.W, Sabesan, M.N, Buehner, M, Ford, G.C, Rossmann, M.G. | | Deposit date: | 1975-07-01 | | Release date: | 1977-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Studies of asymmetry in the three-dimensional structure of lobster D-glyceraldehyde-3-phosphate dehydrogenase.
J.Biol.Chem., 250, 1975
|
|
5IXP
 
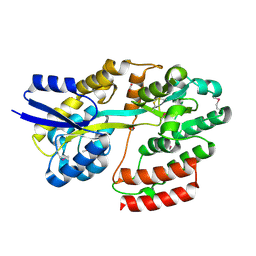 | | Crystal structure of Extracellular solute-binding protein family 1 | | Descriptor: | Extracellular solute-binding protein family 1, FORMIC ACID | | Authors: | Chang, C, Cuff, M, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-03-23 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of Extracellular solute-binding protein family 1
To Be Published
|
|
4UV4
 
 | | Crystal structure of anti-FPR Fpro0165 Fab fragment | | Descriptor: | FPRO0165 FAB | | Authors: | Douthwaite, J.A, Sridharan, S, Huntington, C, Marwood, R, Hammersley, J, Hakulinen, J.K, Ek, M, Sjogren, T, Rider, D, Privezentzev, C, Seaman, J.C, Cariuk, P, Knights, V, Young, J, Wilkinson, T, Sleeman, M, Finch, D.K, Lowe, D.C, Vaughan, T.J. | | Deposit date: | 2014-08-04 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Affinity Maturation of a Novel Antagonistic Human Monoclonal Antibody with a Long Vh Cdr3 Targeting the Class a Gpcr Formyl-Peptide Receptor 1.
Mabs, 7, 2015
|
|
3B7T
 
 | | [E296Q]LTA4H in complex with Arg-Ala-Arg substrate | | Descriptor: | IMIDAZOLE, Leukotriene A-4 hydrolase, RAR peptide, ... | | Authors: | Tholander, F, Haeggstrom, J, Thunnissen, M, Muroya, A, Roques, B.-P, Fournie-Zaluski, M.-C. | | Deposit date: | 2007-10-31 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based dissection of the active site chemistry of leukotriene a4 hydrolase: implications for m1 aminopeptidases and inhibitor design.
Chem.Biol., 15, 2008
|
|
6ZFX
 
 | | hSARM1 GraFix-ed | | Descriptor: | (~{E})-4-methylnon-4-enedial, NAD(+) hydrolase SARM1 | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-18 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
1QOB
 
 | | FERREDOXIN MUTATION D62K | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN, SULFATE ION | | Authors: | Holden, H.M, Benning, M.M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
