4XSA
 
 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase | | Authors: | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
4XS7
 
 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase | | Authors: | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
4XS9
 
 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase, N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-N-[2-(propanoylamino)ethyl]-beta-alaninamide | | Authors: | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
4XSB
 
 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase | | Authors: | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
5TT4
 
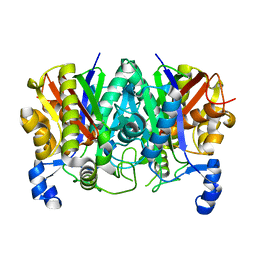 | | Determining the Molecular Basis For Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase | | Authors: | Jackson, D.R, Valentic, T.R, Patel, A, Tsai, S.C, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2016-11-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determining the Molecular Basis For Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
5KOF
 
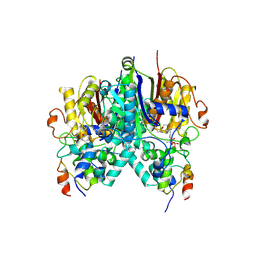 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and Acyl Carrier Protein, AcpP | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 1, Acyl carrier protein, [(3~{S})-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-4-[[3-oxidanylidene-3-[2-(prop-2-enoylamino)ethylamino]propyl]amino]butyl] dihydrogen phosphate | | Authors: | Milligan, J.C, Jackson, D.R, Barajas, J.F, Tsai, S.C. | | Deposit date: | 2016-06-30 | | Release date: | 2018-01-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for interactions between an acyl carrier protein and a ketosynthase.
Nat.Chem.Biol., 15, 2019
|
|
1UUH
 
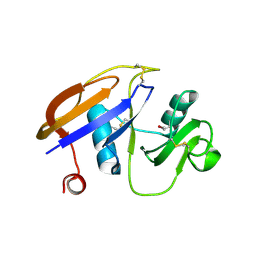 | | Hyaluronan binding domain of human CD44 | | Descriptor: | CD44 ANTIGEN | | Authors: | Teriete, P, Banerji, S, Noble, M, Blundell, C, Wright, A, Pickford, A, Lowe, E, Mahoney, D, Tammi, M, Kahmann, J, Campbell, I, Day, A, Jackson, D. | | Deposit date: | 2003-12-19 | | Release date: | 2004-03-04 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Regulatory Hyaluronan-Binding Domain in the Inflammatory Leukocyte Homing Receptor Cd44
Mol.Cell, 13, 2004
|
|
5KBZ
 
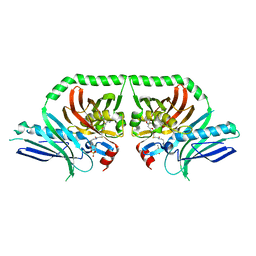 | | Structure of the PksA Product Template domain in complex with a phosphopantetheine mimetic | | Descriptor: | (14R)-14-hydroxy-15,15-dimethyl-1-[5-({[(5-methyl-1,2-oxazol-3-yl)methyl]sulfanyl}methyl)-1,2-oxazol-3-yl]-4,9,13-trioxo-2-thia-5,8,12-triazahexadecan-16-yl dihydrogen phosphate, Noranthrone synthase | | Authors: | Tsai, S.C, Burkart, M.D, Townsend, C.A, Barajas, J.F, Shakya, G, Topper, C.L, Moreno, G, Jackson, D.R, Rivera, H, La Clair, J.J, Vagstad, A. | | Deposit date: | 2016-06-03 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Polyketide mimetics yield structural and mechanistic insights into product template domain function in nonreducing polyketide synthases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1POZ
 
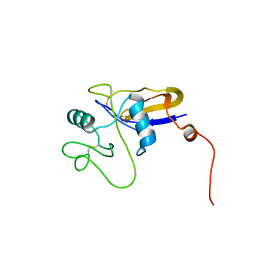 | | SOLUTION STRUCTURE OF THE HYALURONAN BINDING DOMAIN OF HUMAN CD44 | | Descriptor: | CD44 antigen | | Authors: | Teriete, P, Banerji, S, Blundell, C.D, Kahmann, J.D, Pickford, A.R, Wright, A.J, Campbell, I.D, Jackson, D.G, Day, A.J. | | Deposit date: | 2003-06-16 | | Release date: | 2004-03-16 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the Regulatory Hyaluronan Binding Domain in the Inflammatory Leukocyte Homing Receptor CD44.
Mol.Cell, 13, 2004
|
|
2JCQ
 
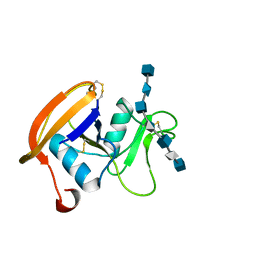 | | The hyaluronan binding domain of murine CD44 in a Type A complex with an HA 8-mer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD44 ANTIGEN, GLYCEROL | | Authors: | Banerji, S, Wright, A.J, Noble, M.E.M, Mahoney, D.J, Campbell, I.D, Day, A.J, Jackson, D.G. | | Deposit date: | 2007-01-03 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structures of the Cd44-Hyaluronan Complex Provide Insight Into a Fundamental Carbohydrate-Protein Interaction.
Nat.Struct.Mol.Biol., 14, 2008
|
|
2JCR
 
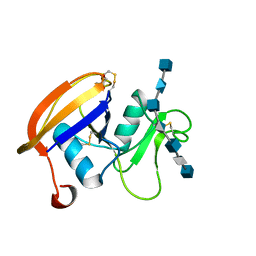 | | The hyaluronan binding domain of murine CD44 in a Type B complex with an HA 8-mer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD44 ANTIGEN, GLYCEROL | | Authors: | Banerji, S, Wright, A.J, Noble, M.E.M, Mahoney, D.J, Campbell, I.D, Day, A.J, Jackson, D.G. | | Deposit date: | 2007-01-03 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Cd44-Hyaluronan Complex Provide Insight Into a Fundamental Carbohydrate-Protein Interaction.
Nat.Struct.Mol.Biol., 14, 2008
|
|
4WV3
 
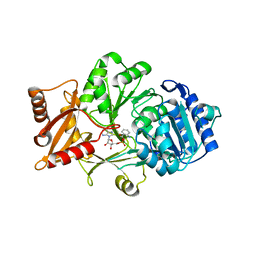 | | Crystal structure of the anthranilate CoA ligase AuaEII in complex with anthranoyl-AMP | | Descriptor: | 5'-O-[(S)-[(2-aminobenzoyl)oxy](hydroxy)phosphoryl]adenosine, Anthranilate-CoA ligase | | Authors: | Tsai, S.C, Muller, R, Jackson, D.R, Tu, S.S, Nguyen, M, Barajas, J.F, Pistorious, D, Krug, D. | | Deposit date: | 2014-11-04 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Structural Insights into Anthranilate Priming during Type II Polyketide Biosynthesis.
Acs Chem.Biol., 11, 2016
|
|
2JCP
 
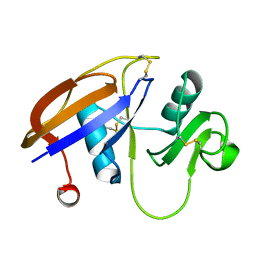 | | The hyaluronan binding domain of murine CD44 | | Descriptor: | CD44 ANTIGEN | | Authors: | Banerji, S, Wright, A.J, Noble, M.E.M, Mahoney, D.J, Campbell, I.D, Day, A.J, Jackson, D.G. | | Deposit date: | 2007-01-03 | | Release date: | 2007-01-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the Cd44-Hyaluronan Complex Provide Insight Into a Fundamental Carbohydrate-Protein Interaction.
Nat.Struct.Mol.Biol., 14, 2008
|
|
1GJG
 
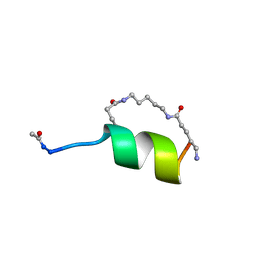 | | Peptide Antagonist of IGFBP1, (i,i+8) Covalently Restrained Analog, Minimized Average Structure | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1GJF
 
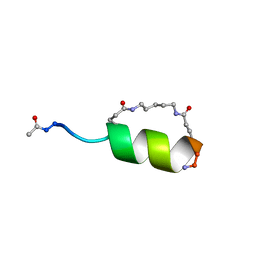 | | Peptide Antagonist of IGFBP1, (i,i+7) Covalently Restrained Analog, Minimized Average Structure | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
4X4J
 
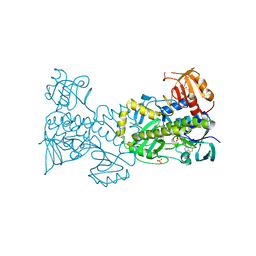 | | Structural and Functional Studies of BexE: Insights into Oxidation During BE-7585A Biosynthesis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative oxygenase, SULFATE ION | | Authors: | Tsai, S.-C, Jackson, D.R, Patel, A, Barajas, J.F, Rohr, J, Yu, X, Liu, H.-W, Sasaki, E, Calveras, J, Metsa-Ketela, M. | | Deposit date: | 2014-12-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and Functional Studies of BexE: Insights into Oxidation During BE-7585A Biosynthesis
To Be Published
|
|
4XRT
 
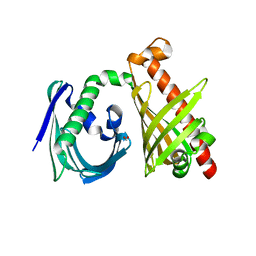 | | Crystal structure of the di-domain ARO/CYC StfQ from the steffimycin biosynthetic pathway | | Descriptor: | FORMIC ACID, StfQ Aromatase/Cyclase | | Authors: | Tsai, S.C, Caldara-Festin, G.M, Jackson, D.R, Aguilar, S, Patel, A, Nguyen, M, Sasaki, E, Valentic, T.R, Barajas, J.F, Vo, M, Khanna, A, Liu, H.-W. | | Deposit date: | 2015-01-21 | | Release date: | 2015-12-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structural and functional analysis of two di-domain aromatase/cyclases from type II polyketide synthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1IN2
 
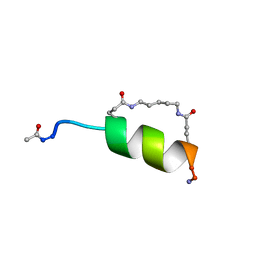 | | Peptide Antagonist of IGFBP1, (i,i+7) Covalently Restrained Analog | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1IN3
 
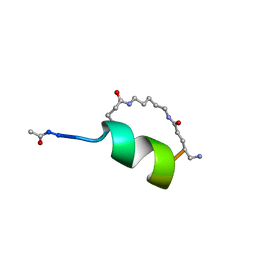 | | Peptide Antagonist of IGFBP1, (i,i+8) Covalently Restrained Analog | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
4XRW
 
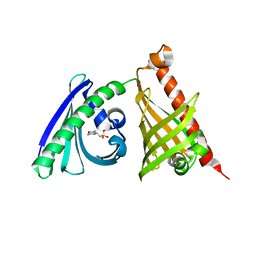 | | Crystal structure of the di-domain ARO/CYC BexL from the BE-7585A biosynthetic pathway | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BexL | | Authors: | Tsai, S.C, Caldara-Festin, G.M, Jackson, D.R, Aguilar, S, Patel, A, Nguyen, M, Sasaki, E, Valentic, T.R, Barajas, J.F, Vo, M, Khanna, A, Liu, H.-W. | | Deposit date: | 2015-01-21 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural and functional analysis of two di-domain aromatase/cyclases from type II polyketide synthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3L4Q
 
 | | Structural insights into phosphoinositide 3-kinase activation by the influenza A virus NS1 protein | | Descriptor: | GLYCEROL, Non-structural protein 1, Phosphatidylinositol 3-kinase regulatory subunit beta | | Authors: | Hale, B.G, Kerry, P.S, Jackson, D, Precious, B.L, Gray, A, Killip, M.J, Randall, R.E, Russell, R.J. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into phosphoinositide 3-kinase activation by the influenza A virus NS1 protein
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KPM
 
 | | Crystal Structure of HLA B*4402 in complex with EEYLKAWTF, a mimotope | | Descriptor: | Beta-2-microglobulin, EEYLKAWTF, mimotope peptide, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
3KPQ
 
 | | Crystal Structure of HLA B*4405 in complex with EEYLKAWTF, a mimotope | | Descriptor: | Beta-2-microglobulin, EEYLKAWTF, mimotope peptide, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
3KPL
 
 | | Crystal Structure of HLA B*4402 in complex with EEYLQAFTY a self peptide from the ABCD3 protein | | Descriptor: | Beta-2-microglobulin, EEYLQAFTY, self peptide from the ATP binding cassette protein ABCD3, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
3KPS
 
 | | Crystal Structure of the LC13 TCR in complex with HLA B*4405 bound to EEYLQAFTY a self peptide from the ABCD3 protein | | Descriptor: | Beta-2-microglobulin, EEYLQAFTY, self peptide from the ATP binding cassette protein ABCD3, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
