4UC0
 
 | | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis | | Descriptor: | HYPOXANTHINE, Purine nucleoside phosphorylase | | Authors: | Cameron, S.A, Sampathkumar, P, Ramagopal, U.A, Attonito, J, Ahmed, M, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Glenn, A.S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Wasserman, S.R, Schramm, V.L, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis
To be published
|
|
7QWV
 
 | | Crystal structure of the REC114-TOPOVIBL complex. | | Descriptor: | Meiotic recombination protein REC114, Type 2 DNA topoisomerase 6 subunit B-like | | Authors: | Juarez-Martinez, A.B, Robert, T, de Massy, B, Kadlec, J. | | Deposit date: | 2022-01-25 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | TOPOVIBL-REC114 interaction regulates meiotic DNA double-strand breaks.
Nat Commun, 13, 2022
|
|
6JUI
 
 | | The atypical Myb-like protein Cdc5 contains two distinct nucleic acid-binding surfaces | | Descriptor: | Pre-mRNA-splicing factor CEF1 | | Authors: | Wang, C, Li, G, Li, M, Yang, J, Liu, J. | | Deposit date: | 2019-04-14 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Two distinct nucleic acid binding surfaces of Cdc5 regulate development.
Biochem.J., 476, 2019
|
|
6DLB
 
 | |
4U7P
 
 | | Crystal structure of DNMT3A-DNMT3L complex | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Wang, L, Guo, X, Li, J, Xiao, J, Yin, X, He, S, Wang, J, Xu, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.821 Å) | | Cite: | Structural insight into autoinhibition and histone H3-induced activation of DNMT3A
Nature, 517, 2015
|
|
4OCZ
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with 1-(1-isobutyrylpiperidin-4-yl)-3-(4-(trifluoromethyl)phenyl)urea | | Descriptor: | 1-[1-(2-methylpropanoyl)piperidin-4-yl]-3-[4-(trifluoromethyl)phenyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Lee, K.S.S, Liu, J, Wagner, K.M, Pakhomova, S, Dong, H, Morriseau, C, Fu, S.H, Yang, J, Wang, P, Ulu, A, Mate, C, Nguyen, L, Wullf, H, Eldin, M.L, Mara, A.A, Newcomer, M.E, Zeldin, D.C, Hammock, B.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy.
J.Med.Chem., 57, 2014
|
|
6DLA
 
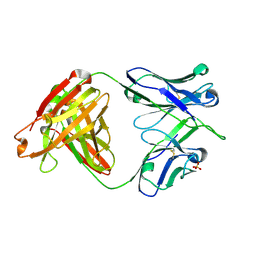 | |
6K9L
 
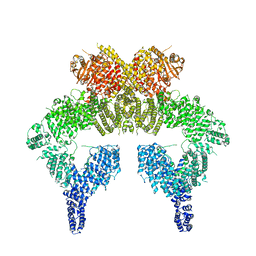 | | 4.27 Angstrom resolution cryo-EM structure of human dimeric ATM kinase | | Descriptor: | Serine-protein kinase ATM | | Authors: | Xiao, J, Liu, M, Qi, Y, Chaban, Y, Gao, C, Tian, Y, Yu, Z, Li, J, Zhang, P, Xu, Y. | | Deposit date: | 2019-06-16 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural insights into the activation of ATM kinase.
Cell Res., 29, 2019
|
|
4FEP
 
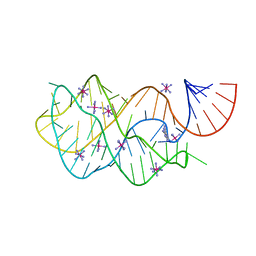 | | Crystal structure of the A24U/U25A/A46G/C74U mutant xpt-pbuX guanine riboswitch aptamer domain in complex with 2,6-diaminopurine | | Descriptor: | 9H-PURINE-2,6-DIAMINE, A24U/U25A/A46G/C74U mutant of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain, COBALT HEXAMMINE(III) | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
7R6H
 
 | | RHCC in complex with o-carborane | | Descriptor: | Tetrabrachion, ortho-carborane | | Authors: | Heide, F, McDougall, M, Stetefeld, J. | | Deposit date: | 2021-06-22 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Boron rich nanotube drug carrier system is suited for boron neutron capture therapy.
Sci Rep, 11, 2021
|
|
6KA4
 
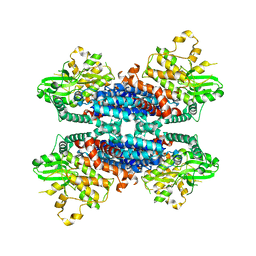 | | Cryo-EM structure of the AtMLKL3 tetramer | | Descriptor: | F22L4.1 protein | | Authors: | Lisa, M, Huang, M, Zhang, X, Ryohei, T.N, Leila, B.K, Isabel, M.L.S, Florence, J, Viera, K, Dmitry, L, Jane, E.P, James, M.M, Kay, H, Paul, S.L, Chai, J, Takaki, M. | | Deposit date: | 2019-06-20 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the AtMLKL3 tetramer
To Be Published
|
|
7REC
 
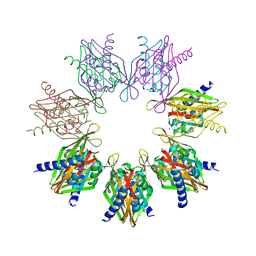 | | Structure of Thr354Asn, Glu355Gln, Thr412Asn, Ile414Met, Ile464His, and Phe467Met mutant human CaMKII alpha hub bound to 5-HDC | | Descriptor: | 5-hydroxydiclofenac, Calcium/calmodulin-dependent protein kinase type II subunit alpha, SODIUM ION | | Authors: | McSpadden, E.D, Chi, C.C, Gee, C.L, Kuriyan, J. | | Deposit date: | 2021-07-12 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | GHB analogs confer neuroprotection through specific interaction with the CaMKII alpha hub domain.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6D6Z
 
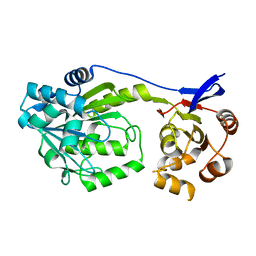 | |
6D8B
 
 | |
4FEO
 
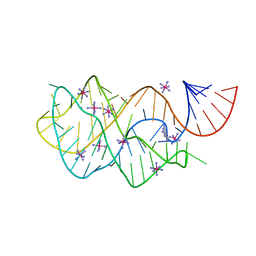 | | Crystal structure of the AU25A/A46G/C74U mutant xpt-pbuX guanine riboswitch aptamer domain in complex with 2,6-diaminopurine | | Descriptor: | 9H-PURINE-2,6-DIAMINE, COBALT HEXAMMINE(III), U25A/A46G/C74U mutant of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
7XRY
 
 | | Crystal structure of MERS main protease in complex with inhibitor YH-53 | | Descriptor: | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, ORF1a | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2022-05-12 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
7XRS
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with inhibitor YH-53 | | Descriptor: | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, Replicase polyprotein 1a | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-05-11 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
6DDT
 
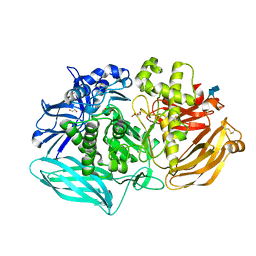 | | mouse beta-mannosidase (MANBA) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gytz, H, Liang, J, Liang, Y, Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2018-05-10 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of mammalian beta-mannosidase provides insight into beta-mannosidosis and nystagmus.
FEBS J., 286, 2019
|
|
4TWE
 
 | | Structure of ligand-free N-acetylated-alpha-linked-acidic-dipeptidase like protein (NAALADaseL) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tykvart, J, Barinka, C, Lubkowski, J, Sacha, P, Konvalinka, J. | | Deposit date: | 2014-06-30 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical characterization of a novel aminopeptidase from human intestine.
J.Biol.Chem., 290, 2015
|
|
7YGQ
 
 | | Crystal structure of SARS main protease in complex with inhibitor YH-53 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-07-12 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
7Y5N
 
 | | Structure of 1:1 PAPP-A.ProMBP complex(half map) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bone marrow proteoglycan, ... | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-06-17 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
7Y5Q
 
 | | Structure of 1:1 PAPP-A.STC2 complex(half map) | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Pappalysin-1, Stanniocalcin-2, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-06-17 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
7YIM
 
 | | Cryo-EM structure of human Alpha-fetoprotein | | Descriptor: | Alpha-fetoprotein | | Authors: | Liu, N, Liu, K, Wu, C, Liu, Z, Li, M, Wang, J, Wang, H.W. | | Deposit date: | 2022-07-17 | | Release date: | 2023-01-18 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Uniform thin ice on ultraflat graphene for high-resolution cryo-EM.
Nat.Methods, 20, 2023
|
|
6DCJ
 
 | |
6L6W
 
 | | The structure of ScoE with intermediate | | Descriptor: | (3R)-3-[[(1R)-1,2-bis(oxidanyl)-2-oxidanylidene-ethyl]amino]butanoic acid, FE (II) ION, FORMIC ACID, ... | | Authors: | Chen, T.Y, Chen, J, Zhou, J, Chang, W. | | Deposit date: | 2019-10-29 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Pathway from N-Alkylglycine to Alkylisonitrile Catalyzed by Iron(II) and 2-Oxoglutarate-Dependent Oxygenases.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
