1XY1
 
 | | CRYSTAL STRUCTURE ANALYSIS OF DEAMINO-OXYTOCIN. CONFORMATIONAL FLEXIBILITY AND RECEPTOR BINDING | | Descriptor: | BETA-MERCAPTOPROPIONATE-OXYTOCIN | | Authors: | Husain, J, Blundell, T.L, Wood, S.P, Tickle, I.J, Cooper, S, Pitts, J.E. | | Deposit date: | 1987-06-05 | | Release date: | 1988-04-16 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystal structure analysis of deamino-oxytocin: conformational flexibility and receptor binding.
Science, 232, 1986
|
|
5HE0
 
 | | Bovine GRK2 in complex with Gbetagamma subunits and CCG215022 | | Descriptor: | (4S)-4-{4-fluoro-3-[(pyridin-2-ylmethyl)carbamoyl]phenyl}-N-(1H-indazol-5-yl)-6-methyl-2-oxo-1,2,3,4-tetrahydropyrimidine-5-carboxamide, Beta-adrenergic receptor kinase 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cato, M.C, Waninger-Saroni, J, Tesmer, J.J.G. | | Deposit date: | 2016-01-05 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Highly Selective and Potent G Protein-Coupled Receptor Kinase 2 Inhibitors.
J.Med.Chem., 59, 2016
|
|
1RP5
 
 | | PBP2x from Streptococcus pneumoniae strain 5259 with reduced susceptibility to beta-lactam antibiotics | | Descriptor: | SULFATE ION, penicillin-binding protein 2x | | Authors: | Pernot, L, Chesnel, L, Legouellec, A, Croize, J, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2003-12-03 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A PBP2x from a clinical isolate of Streptococcus pneumoniae exhibits an alternative mechanism for reduction of susceptibility to beta-lactam antibiotics.
J.Biol.Chem., 279, 2004
|
|
5H5Y
 
 | | Structure of Transferase mutant-C23S,C199S | | Descriptor: | Non-LEE encoded effector protein NleB | | Authors: | Park, J.B, Yoo, Y, Kim, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
1RQQ
 
 | | Crystal Structure of the Insulin Receptor Kinase in Complex with the SH2 Domain of APS | | Descriptor: | BISUBSTRATE INHIBITOR, Insulin receptor, MANGANESE (II) ION, ... | | Authors: | Hu, J, Liu, J, Ghirlando, R, Saltiel, A.R, Hubbard, S.R. | | Deposit date: | 2003-12-06 | | Release date: | 2003-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for recruitment of the adaptor protein APS to the activated insulin receptor.
Mol.Cell, 12, 2003
|
|
4KAN
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 2-(2,5-dimethyl-1,3-thiazol-4-yl)acetic acid | | Descriptor: | (2,5-dimethyl-1,3-thiazol-4-yl)acetic acid, ADENOSINE, POTASSIUM ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-22 | | Release date: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 2-(2,5-dimethyl-1,3-thiazol-4-yl)acetic acid
To be Published
|
|
5H6B
 
 | | Crystal structure of a thermostable lipase from Marine Streptomyces | | Descriptor: | ACETATE ION, IMIDAZOLE, Putative secreted lipase, ... | | Authors: | Hou, S, Zhao, Z, Liu, J. | | Deposit date: | 2016-11-11 | | Release date: | 2017-09-20 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a lipase from Streptomyces sp. strain W007 - implications for thermostability and regiospecificity
FEBS J., 284, 2017
|
|
4PRA
 
 | | Crystal structure of a HLA-B*35:01-HPVG-Q5 | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 1, MHC class I antigen | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
4PU4
 
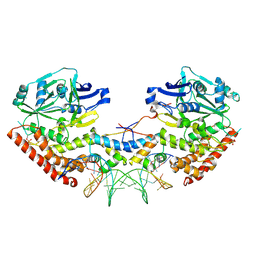 | | Shewanella oneidensis MR-1 Toxin Antitoxin System HipA, HipB and its operator DNA complex (space group P21) | | Descriptor: | Operator DNA, Toxin-antitoxin system antidote transcriptional repressor Xre family, Toxin-antitoxin system toxin HipA family | | Authors: | Wen, Y, Behiels, E, Felix, J, Elegheert, J, Vergauwen, B, Devreese, B, Savvides, S. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.786 Å) | | Cite: | The bacterial antitoxin HipB establishes a ternary complex with operator DNA and phosphorylated toxin HipA to regulate bacterial persistence.
Nucleic Acids Res., 42, 2014
|
|
1RML
 
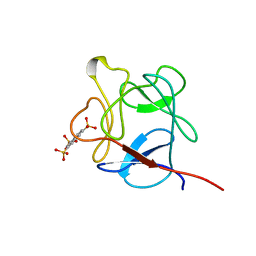 | | NMR STUDY OF ACID FIBROBLAST GROWTH FACTOR BOUND TO 1,3,6-NAPHTHALENE TRISULPHONATE, 26 STRUCTURES | | Descriptor: | ACIDIC FIBROBLAST GROWTH FACTOR, NAPHTHALENE TRISULFONATE | | Authors: | Lozano, R.M, Jimenez, M.A, Santoro, J, Rico, M, Gimenez-Gallego, G. | | Deposit date: | 1998-05-21 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of acidic fibroblast growth factor bound to 1,3, 6-naphthalenetrisulfonate: a minimal model for the anti-tumoral action of suramins and suradistas.
J.Mol.Biol., 281, 1998
|
|
2O52
 
 | | Crystal structure of human RAB4B in complex with GDP | | Descriptor: | BETA-MERCAPTOETHANOL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhu, H, Tempel, W, Wang, J, Shen, Y, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-05 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human RAB4B in complex with GDP
To be Published
|
|
4PQW
 
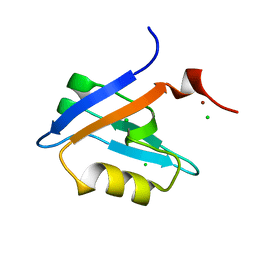 | | Crystal Structure of Phospholipase C beta 3 in Complex with PDZ1 of NHERF1 | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Jiang, Y, Wang, S, Holcomb, J, Trescott, L, Guan, X, Hou, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallographic analysis of NHERF1-PLC beta 3 interaction provides structural basis for CXCR2 signaling in pancreatic cancer.
Biochem.Biophys.Res.Commun., 446, 2014
|
|
5WZ1
 
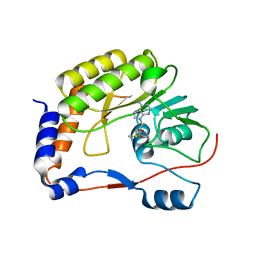 | | Crystal structure of Zika virus NS5 methyltransferase bound to S-adenosyl-L-methionine | | Descriptor: | NS5 methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Duan, W, Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | The crystal structure of Zika virus NS5 reveals conserved drug targets.
EMBO J., 36, 2017
|
|
7LJY
 
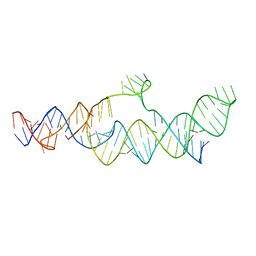 | | Cryo-EM structure of the B dENE construct complexed with a 28-mer poly(A) | | Descriptor: | B dENE construct, poly(A) | | Authors: | Torabi, S, Chen, Y, Zhang, K, Wang, J, DeGregorio, S, Vaidya, A, Su, Z, Pabit, S, Chiu, W, Pollack, L, Steitz, J. | | Deposit date: | 2021-02-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural analyses of an RNA stability element interacting with poly(A).
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5G02
 
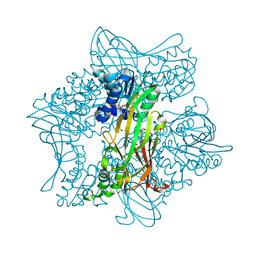 | | Crystal Structure of zebrafish Protein Arginine Methyltransferase 2 with SFG | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, LITHIUM ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
FEBS J., 284, 2017
|
|
4PU7
 
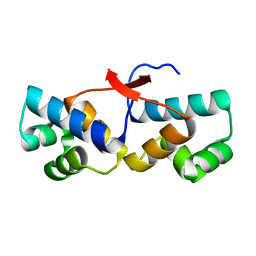 | | Shewanella oneidensis Toxin Antitoxin System Antitoxin Protein HipB Resolution 1.85 | | Descriptor: | Toxin-antitoxin system antidote transcriptional repressor Xre family | | Authors: | Wen, Y, Behiels, E, Felix, J, Elegheert, J, Vergauwen, B, Devreese, B, Savvides, S. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The bacterial antitoxin HipB establishes a ternary complex with operator DNA and phosphorylated toxin HipA to regulate bacterial persistence.
Nucleic Acids Res., 42, 2014
|
|
5WZH
 
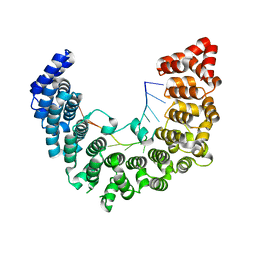 | | Structure of APUM23-GGAAUUGACGG | | Descriptor: | Pumilio homolog 23, RNA (5'-R(*GP*GP*AP*AP*UP*UP*GP*AP*CP*GP*G)-3') | | Authors: | Bao, H, Wang, N, Wang, C, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-01-18 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Structural basis for the specific recognition of 18S rRNA by APUM23.
Nucleic Acids Res., 45, 2017
|
|
4PUA
 
 | | Crystal Structure Of glutathione transferase YghU from Streptococcus pneumoniae ATCC 700669, complexed with glutathione, Target EFI-507284 | | Descriptor: | GLUTATHIONE, glutathione S-transferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-12 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Crystal Structure Of glutathione transferase YghU from Streptococcus pneumoniae ATCC 700669, complexed with glutathione, Target EFI-507284
To be Published
|
|
1R67
 
 | | Y104A MUTANT OF E.COLI IPP ISOMERASE | | Descriptor: | Isopentenyl-diphosphate delta-isomerase, MAGNESIUM ION | | Authors: | Wouters, J. | | Deposit date: | 2003-10-15 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural role for Tyr-104 in Escherichia coli isopentenyl-diphosphate isomerase: site-directed mutagenesis, enzymology, and protein crystallography.
J.Biol.Chem., 281, 2006
|
|
1HH6
 
 | | ANTI-P24 (HIV-1) FAB FRAGMENT CB41 COMPLEXED WITH A PEPTIDE | | Descriptor: | IGG2A KAPPA ANTIBODY CB41 (HEAVY CHAIN), IGG2A KAPPA ANTIBODY CB41 (LIGHT CHAIN), PEP-4 | | Authors: | Hahn, M, Wessner, H, Schneider-Mergener, J, Hohne, W. | | Deposit date: | 2000-12-21 | | Release date: | 2001-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolutionary Transition Pathways for Changing Peptide Ligand Specificity and Structure
Embo J., 19, 2000
|
|
4PRH
 
 | | Crystal structure of TK3 TCR-HLA-B*35:08-HPVG-D5 complex | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 1, MHC class I antigen, ... | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
1ZGL
 
 | | Crystal structure of 3A6 TCR bound to MBP/HLA-DR2a | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, Myelin basic protein, ... | | Authors: | Li, Y, Huang, Y, Lue, J, Quandt, J.A, Martin, R, Mariuzza, R.A. | | Deposit date: | 2005-04-21 | | Release date: | 2005-10-18 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule.
Embo J., 24, 2005
|
|
6K9K
 
 | | Monomeric human ATM (Ataxia telangiectasia mutated) kinase | | Descriptor: | Serine-protein kinase ATM | | Authors: | Xiao, J, Liu, M, Qi, Y, Yuriy, C, Zhang, P, Xu, Y. | | Deposit date: | 2019-06-16 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.82 Å) | | Cite: | Structural insights into the activation of ATM kinase.
Cell Res., 29, 2019
|
|
5FOB
 
 | | Crystal Structure of Human Complement C3b in complex with Smallpox Inhibitor of Complement (SPICE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COMPLEMENT C3 BETA CHAIN, ... | | Authors: | Forneris, F, Wu, J, Xue, X, Gros, P. | | Deposit date: | 2015-11-18 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
6O0H
 
 | |
