122D
 
 | |
123D
 
 | |
2AYH
 
 | | CRYSTAL AND MOLECULAR STRUCTURE AT 1.6 ANGSTROMS RESOLUTION OF THE HYBRID BACILLUS ENDO-1,3-1,4-BETA-D-GLUCAN 4-GLUCANOHYDROLASE H(A16-M) | | Descriptor: | 1,3-1,4-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, CALCIUM ION | | Authors: | Hahn, M, Keitel, T, Heinemann, U. | | Deposit date: | 1995-02-02 | | Release date: | 1995-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal and molecular structure at 0.16-nm resolution of the hybrid Bacillus endo-1,3-1,4-beta-D-glucan 4-glucanohydrolase H(A16-M).
Eur.J.Biochem., 232, 1995
|
|
1YMM
 
 | | TCR/HLA-DR2b/MBP-peptide complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Hahn, M, Nicholson, M.J, Pyrdol, J, Wucherpfennig, K.W. | | Deposit date: | 2005-01-21 | | Release date: | 2005-05-03 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Unconventional topology of self peptide-major histocompatibility complex binding by a human autoimmune T cell receptor.
NAT.IMMUNOL., 6, 2005
|
|
1HH9
 
 | | ANTI-P24 (HIV-1) FAB FRAGMENT CB41 COMPLEXED WITH A PEPTIDE | | Descriptor: | IGG2A KAPPA ANTIBODY CB41 (HEAVY CHAIN), IGG2A KAPPA ANTIBODY CB41 (LIGHT CHAIN), PEP-2 | | Authors: | Hahn, M, Wessner, H, Schneider-Mergener, J, Hohne, W. | | Deposit date: | 2000-12-21 | | Release date: | 2001-01-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Evolutionary Transition Pathways for Changing Peptide Ligand Specificity and Structure
Embo J., 19, 2000
|
|
1HH6
 
 | | ANTI-P24 (HIV-1) FAB FRAGMENT CB41 COMPLEXED WITH A PEPTIDE | | Descriptor: | IGG2A KAPPA ANTIBODY CB41 (HEAVY CHAIN), IGG2A KAPPA ANTIBODY CB41 (LIGHT CHAIN), PEP-4 | | Authors: | Hahn, M, Wessner, H, Schneider-Mergener, J, Hohne, W. | | Deposit date: | 2000-12-21 | | Release date: | 2001-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolutionary Transition Pathways for Changing Peptide Ligand Specificity and Structure
Embo J., 19, 2000
|
|
1DXJ
 
 | | Structure of the chitinase from jack bean | | Descriptor: | CLASS II CHITINASE, SULFATE ION | | Authors: | Hahn, M, Hennig, M, Schlesier, B, Hohne, W. | | Deposit date: | 2000-01-10 | | Release date: | 2000-08-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Jack Bean Chitinase
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1E4X
 
 | | crossreactive binding of a circularized peptide to an anti-TGFalpha antibody Fab-fragment | | Descriptor: | CYCLIC PEPTIDE, TAB2 | | Authors: | Hahn, M, Winkler, D, Misselwitz, R, Wessner, H, Welfle, K, Zahn, G, Schneider-Mergener, J, Hoehne, W. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2017-02-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-Reactive Binding of Cyclic Peptides to an Anti-Tgf Alpha Antibody Fab Fragment: An X-Ray Structural and Thermodynamic Analysis
J.Mol.Biol., 314, 2001
|
|
1E4W
 
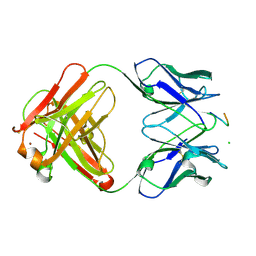 | | crossreactive binding of a circularized peptide to an anti-TGFalpha antibody Fab-fragment | | Descriptor: | CHLORIDE ION, CYCLIC PEPTIDE, NICKEL (II) ION, ... | | Authors: | Hahn, M, Winkler, D, Misselwitz, R, Wessner, H, Welfle, K, Zahn, G, Schneider-Mergener, J, Hoehne, W. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cross-Reactive Binding of Cyclic Peptides to an Anti-Tgf Alpha Antibody Fab Fragment: An X-Ray Structural and Thermodynamic Analysis
J.Mol.Biol., 314, 2001
|
|
1MAC
 
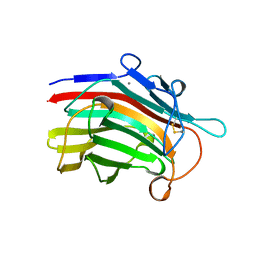 | | CRYSTAL STRUCTURE AND SITE-DIRECTED MUTAGENESIS OF BACILLUS MACERANS ENDO-1,3-1,4-BETA-GLUCANASE | | Descriptor: | 1,3-1,4-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, CALCIUM ION | | Authors: | Hahn, M, Heinemann, U. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and site-directed mutagenesis of Bacillus macerans endo-1,3-1,4-beta-glucanase.
J.Biol.Chem., 270, 1995
|
|
1CPN
 
 | |
1CPM
 
 | |
1GBG
 
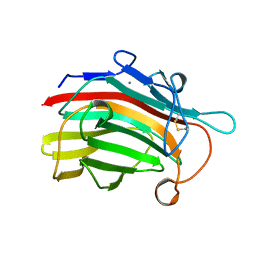 | | BACILLUS LICHENIFORMIS BETA-GLUCANASE | | Descriptor: | (1,3-1,4)-BETA-D-GLUCAN 4 GLUCANOHYDROLASE, CALCIUM ION | | Authors: | Hahn, M, Heinemann, U. | | Deposit date: | 1995-08-25 | | Release date: | 1995-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus licheniformis 1,3-1,4-beta-D-glucan 4-glucanohydrolase at 1.8 A resolution.
FEBS Lett., 374, 1995
|
|
1HI6
 
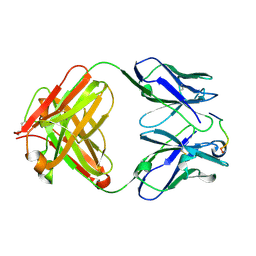 | | ANTI-P24 (HIV-1) FAB FRAGMENT CB41 COMPLEXED WITH A PEPTIDE | | Descriptor: | IGG2A KAPPA ANTIBODY CB41 (HEAVY CHAIN), IGG2A KAPPA ANTIBODY CB41 (LIGHT CHAIN), PEPTIDE 5 | | Authors: | Hahn, M, Wessner, H, Schneider-Mergener, J, Hohne, W. | | Deposit date: | 2001-01-02 | | Release date: | 2001-02-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystallographic Analysis of Anti-P24 (HIV-1) Monoclonal Antibody Cross-Reactivity and Polyspecificity
Cell(Cambridge,Mass.), 91, 1997
|
|
1JHN
 
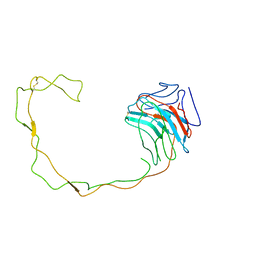 | | Crystal Structure of the Lumenal Domain of Calnexin | | Descriptor: | CALCIUM ION, calnexin | | Authors: | Schrag, J.D, Bergeron, J.M, Li, Y, Borisova, S, Hahn, M, Thomas, D.Y, Cygler, M. | | Deposit date: | 2001-06-28 | | Release date: | 2001-10-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of calnexin, an ER chaperone involved in quality control of protein folding.
Mol.Cell, 8, 2001
|
|
3BG4
 
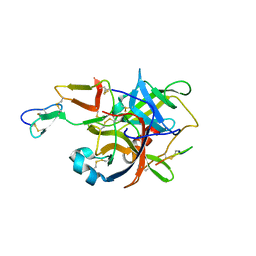 | | The crystal structure of guamerin in complex with chymotrypsin and the development of an elastase-specific inhibitor | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Kim, H, Chu, T.T.T, Kim, D.Y, Kim, D.R, Nguyen, C.M.T, Choi, J, Lee, J.R, Hahn, M.J, Kim, K.K. | | Deposit date: | 2007-11-26 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of guamerin in complex with chymotrypsin and the development of an elastase-specific inhibitor.
J.Mol.Biol., 376, 2008
|
|
1QNJ
 
 | | THE STRUCTURE OF NATIVE PORCINE PANCREATIC ELASTASE AT ATOMIC RESOLUTION (1.1 A) | | Descriptor: | ELASTASE, SODIUM ION, SULFATE ION | | Authors: | Wurtele, M, Hahn, M, Hilpert, K, Hohne, W. | | Deposit date: | 1999-10-15 | | Release date: | 2000-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Structure of Native Porcine Pancreatic Elastase at 1.1 A
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2D25
 
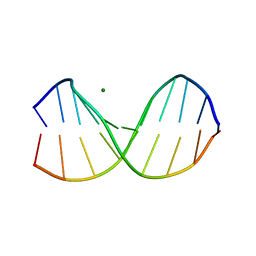 | | C-C-A-G-G-C-M5C-T-G-G; HELICAL FINE STRUCTURE, HYDRATION, AND COMPARISON WITH C-C-A-G-G-C-C-T-G-G | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*GP*CP*(5CM)P*TP*GP*G)-3'), MAGNESIUM ION | | Authors: | Heinemann, U, Hahn, M. | | Deposit date: | 1991-04-23 | | Release date: | 1991-04-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | C-C-A-G-G-C-m5C-T-G-G. Helical fine structure, hydration, and comparison with C-C-A-G-G-C-C-T-G-G.
J.Biol.Chem., 267, 1992
|
|
1DZB
 
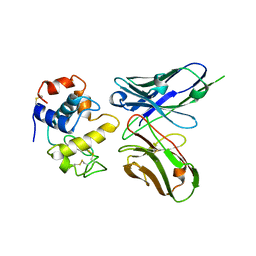 | | Crystal structure of phage library-derived single-chain Fv fragment 1F9 in complex with turkey egg-white lysozyme | | Descriptor: | SCFV FRAGMENT 1F9, TURKEY EGG-WHITE LYSOZYME C | | Authors: | Ay, J, Keitel, T, Kuettner, G, Wessner, H, Scholz, C, Hahn, M, Hoehne, W. | | Deposit date: | 2000-02-23 | | Release date: | 2000-11-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Phage Library-Derived Single-Chain Fv Fragment Complexed with Turkey Egg -White Lysozyme at 2.0 A Resolution
J.Mol.Biol., 301, 2000
|
|
