8GRA
 
 | |
8H3X
 
 | | Bacteroide Fragilis Toxin in complex with nanobody 282 | | Descriptor: | Fragilysin, ZINC ION, nanobody 282 | | Authors: | Wen, Y, Guo, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Screening and epitope characterization of diagnostic nanobody against total and activated Bacteroides fragilis toxin.
Front Immunol, 14, 2023
|
|
6IS1
 
 | |
8H3Y
 
 | | Bacteroide Fragilis Toxin in complex with nanobody 327 | | Descriptor: | Fragilysin, Nanobody 327, ZINC ION | | Authors: | Wen, Y, Guo, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Screening and epitope characterization of diagnostic nanobody against total and activated Bacteroides fragilis toxin.
Front Immunol, 14, 2023
|
|
6IS2
 
 | |
6IS3
 
 | |
6IS4
 
 | |
8GPM
 
 | | Acinetobacter baumannii carbonic anhydrase | | Descriptor: | CALCIUM ION, Carbonic anhydrase, GLYCEROL, ... | | Authors: | Wen, Y, Jiao, M. | | Deposit date: | 2022-08-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Acinetobacter baumannii carbonic anhydrase
Structure, 2023
|
|
7EYM
 
 | | Crystal structure of Vibrio cholerae ppnP | | Descriptor: | Pyrimidine/purine nucleoside phosphorylase | | Authors: | Wen, Y, Wu, B.X. | | Deposit date: | 2021-05-31 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structures of a new class of pyrimidine/purine nucleoside phosphorylase revealed a Cupin fold.
Proteins, 90, 2022
|
|
7EYP
 
 | | Crystal structure of Pseudomonas aeruginosa ppnP | | Descriptor: | Pyrimidine/purine nucleoside phosphorylase | | Authors: | Wen, Y, Wu, B.X. | | Deposit date: | 2021-05-31 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of a new class of pyrimidine/purine nucleoside phosphorylase revealed a Cupin fold.
Proteins, 90, 2022
|
|
7EYK
 
 | |
7EYJ
 
 | | Crystal structure of Escherichia coli ppnP | | Descriptor: | Pyrimidine/purine nucleoside phosphorylase, SULFATE ION | | Authors: | Wen, Y, Wu, B.X. | | Deposit date: | 2021-05-31 | | Release date: | 2022-02-09 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structures of a new class of pyrimidine/purine nucleoside phosphorylase revealed a Cupin fold.
Proteins, 90, 2022
|
|
2MUU
 
 | |
7VCM
 
 | | crystal structure of GINKO1 | | Descriptor: | Green fluorescent protein,Potassium binding protein Kbp,Green fluorescent protein, POTASSIUM ION | | Authors: | Wen, Y, Campbell, R.E, Lemieux, M.J. | | Deposit date: | 2021-09-03 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A sensitive and specific genetically-encoded potassium ion biosensor for in vivo applications across the tree of life.
Plos Biol., 20, 2022
|
|
5IML
 
 | | Nanobody targeting human Vsig4 in Spacegroup P212121 | | Descriptor: | Nanobody, V-set and immunoglobulin domain-containing protein 4 | | Authors: | Wen, Y. | | Deposit date: | 2016-03-06 | | Release date: | 2017-01-11 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evaluation of a nanobody targeting complement receptor Vsig4 and its cross reactivity
Immunobiology, 222, 2017
|
|
5IMK
 
 | | Nanobody targeting human Vsig4 in Spacegroup C2 | | Descriptor: | Nanobody, V-set and immunoglobulin domain-containing protein 4 | | Authors: | Wen, Y. | | Deposit date: | 2016-03-06 | | Release date: | 2017-01-11 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.227 Å) | | Cite: | Structural evaluation of a nanobody targeting complement receptor Vsig4 and its cross reactivity
Immunobiology, 222, 2017
|
|
5IMO
 
 | | Nanobody targeting mouse Vsig4 in Spacegroup P3221 | | Descriptor: | Nanobody, Protein Vsig4 | | Authors: | Wen, Y, Zheng, F. | | Deposit date: | 2016-03-06 | | Release date: | 2017-01-11 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural evaluation of a nanobody targeting complement receptor Vsig4 and its cross reactivity
Immunobiology, 222, 2017
|
|
5IMM
 
 | | Nanobody targeting mouse Vsig4 in Spacegroup P212121 | | Descriptor: | Nanobody, Protein Vsig4 | | Authors: | Wen, Y, Zheng, F. | | Deposit date: | 2016-03-06 | | Release date: | 2017-01-11 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural evaluation of a nanobody targeting complement receptor Vsig4 and its cross reactivity
Immunobiology, 222, 2017
|
|
7EYL
 
 | | Crystal structure of Salmonella enterica ppnP | | Descriptor: | Pyrimidine/purine nucleoside phosphorylase | | Authors: | Wen, Y, Wu, B.X. | | Deposit date: | 2021-05-31 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of a new class of pyrimidine/purine nucleoside phosphorylase revealed a Cupin fold.
Proteins, 90, 2022
|
|
8GPP
 
 | |
6LNP
 
 | | Crystal structure of citrate Biosensor | | Descriptor: | CITRIC ACID, Fusion protein of Green fluorescent protein and Sensor histidine kinase CitA | | Authors: | Wen, Y, Campbell, R. | | Deposit date: | 2019-12-31 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | High-Performance Intensiometric Direct- and Inverse-Response Genetically Encoded Biosensors for Citrate.
Acs Cent.Sci., 6, 2020
|
|
7YW0
 
 | | Bacteroides fragilis Hcp5 | | Descriptor: | Bacterodales T6SS protein TssD (Hcp) | | Authors: | Wen, Y, He, W, Bai, Y. | | Deposit date: | 2022-08-20 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and assembly of type VI secretion system cargo delivery vehicle.
Cell Rep, 42, 2023
|
|
5X5L
 
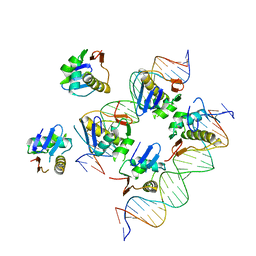 | | Crystal structure of response regulator AdeR DNA binding domain in complex with an intercistronic region | | Descriptor: | AdeR, DNA (5'-D(P*TP*AP*AP*AP*GP*TP*GP*TP*GP*GP*AP*GP*TP*AP*AP*GP*TP*GP*TP*GP*GP*AP*GP*A)-3'), DNA (5'-D(P*TP*CP*TP*CP*CP*AP*CP*AP*CP*TP*TP*AP*CP*TP*CP*CP*AP*CP*AP*CP*TP*TP*TP*A)-3') | | Authors: | Wen, Y. | | Deposit date: | 2017-02-16 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanistic insight into how multidrug resistant Acinetobacter baumannii response regulator AdeR recognizes an intercistronic region.
Nucleic Acids Res., 45, 2017
|
|
5X5J
 
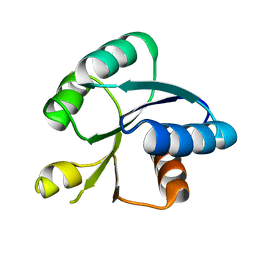 | | Crystal structure of response regulator AdeR receiver domain | | Descriptor: | AdeR | | Authors: | Wen, Y. | | Deposit date: | 2017-02-16 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Mechanistic insight into how multidrug resistant Acinetobacter baumannii response regulator AdeR recognizes an intercistronic region.
Nucleic Acids Res., 45, 2017
|
|
5XJP
 
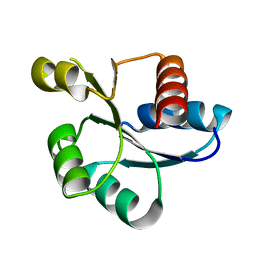 | | Crystal structure of response regulator AdeR receiver domain with Mg | | Descriptor: | AdeR, MAGNESIUM ION | | Authors: | Wen, Y. | | Deposit date: | 2017-05-03 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Mechanistic insight into how multidrug resistant Acinetobacter baumannii response regulator AdeR recognizes an intercistronic region.
Nucleic Acids Res., 45, 2017
|
|
