6A8G
 
 | | The crystal structure of muPAin-1-IG in complex with muPA-SPD at pH8.5 | | Descriptor: | PHOSPHATE ION, Urokinase-type plasminogen activator chain B, muPAin-1-IG | | Authors: | Wang, D, Yang, Y.S, Jiang, L.G, Huang, M.D, Li, J.Y, Andreasen, P.A, Xu, P, Chen, Z. | | Deposit date: | 2018-07-08 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Suppression of Tumor Growth and Metastases by Targeted Intervention in Urokinase Activity with Cyclic Peptides.
J.Med.Chem., 62, 2019
|
|
7CJ1
 
 | |
2OQA
 
 | | X-ray Sequence and Crystal Structure of Luffaculin 1, a Novel Type 1 Ribosome-inactivating Protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Luffaculin 1, ... | | Authors: | Hou, X, Huang, M. | | Deposit date: | 2007-01-31 | | Release date: | 2007-05-29 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray sequence and crystal structure of luffaculin 1, a novel type 1 ribosome-inactivating protein
Bmc Struct.Biol., 7, 2007
|
|
7VM4
 
 | | Crystal structure of uPA in complex with nafamostat | | Descriptor: | 4-carbamimidamidobenzoic acid, TRIETHYLENE GLYCOL, Urokinase-type plasminogen activator | | Authors: | Jiang, L.G, Huang, M.D. | | Deposit date: | 2021-10-07 | | Release date: | 2022-10-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural study of the uPA-nafamostat complex reveals a covalent inhibitory mechanism of nafamostat.
Biophys.J., 121, 2022
|
|
7VM5
 
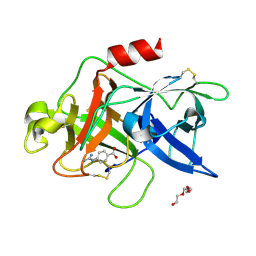 | |
7VM6
 
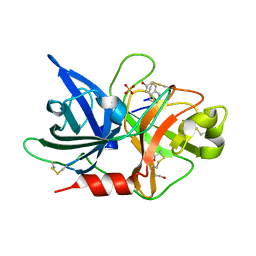 | | Crystal structure of uPA in complex with 6-amidino-2-naphthol | | Descriptor: | 6-oxidanylnaphthalene-2-carboximidamide, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Jiang, L.G, Huang, M.D. | | Deposit date: | 2021-10-07 | | Release date: | 2022-10-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural study of the uPA-nafamostat complex reveals a covalent inhibitory mechanism of nafamostat.
Biophys.J., 121, 2022
|
|
7VM7
 
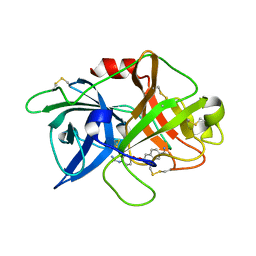 | | Crystal structure of inactive uPA in complex with nafamostat | | Descriptor: | (6-carbamimidoylnaphthalen-2-yl) 4-carbamimidamidobenzoate, Urokinase-type plasminogen activator chain B | | Authors: | Jiang, L.G, Huang, M.D. | | Deposit date: | 2021-10-07 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural study of the uPA-nafamostat complex reveals a covalent inhibitory mechanism of nafamostat.
Biophys.J., 121, 2022
|
|
6AG3
 
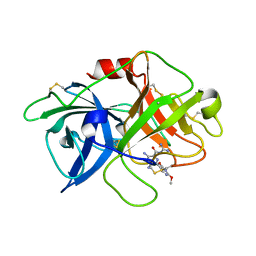 | | Crystal structure of uPA in complex with 3,5-bis(azanyl)-N-carbamimidoyl-6-(2,4-dimethoxypyrimidin-5-yl)pyrazine-2-carboxamide | | Descriptor: | 3,5-bis(azanyl)-N-carbamimidoyl-6-(2,4-dimethoxypyrimidin-5-yl)pyrazine-2-carboxamide, Urokinase-type plasminogen activator | | Authors: | Buckley, B, Jiang, L.G, Majed, H, Huang, M.D, Kelso, M, Ranson, M. | | Deposit date: | 2018-08-09 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | uPA-HMA
To Be Published
|
|
6AEX
 
 | | Crystal structure of unoccupied murine uPAR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor | | Authors: | Min, L, Huang, M. | | Deposit date: | 2018-08-06 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Crystal structure of the unoccupied murine urokinase-type plasminogen activator receptor (uPAR) reveals a tightly packed DII-DIII unit.
Febs Lett., 593, 2019
|
|
6A8O
 
 | | Crystal structures of the serine protease domain of murine plasma kallikrein with peptide inhibitor mupain-1-16 | | Descriptor: | Plasma kallikrein, alpha-D-mannopyranose, peptide inhibitor,, ... | | Authors: | Xu, M, Jiang, L, Huang, M. | | Deposit date: | 2018-07-09 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal structure of plasma kallikrein reveals the unusual flexibility of the S1 pocket triggered by Glu217.
Febs Lett., 592, 2018
|
|
6AG9
 
 | | Crystal structure of uPA in complex with 3,5-bis(azanyl)-6-(1-benzofuran-2-yl)-N-carbamimidoyl-pyrazine-2- carboxamide | | Descriptor: | 3,5-bis(azanyl)-6-(1-benzofuran-2-yl)-N-carbamimidoyl-pyrazine-2-carboxamide, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Buckley, B, Jiang, L.G, Majed, H, Huang, M.D, Kelso, M, Ranson, M. | | Deposit date: | 2018-08-09 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | uPA-HMA
To Be Published
|
|
6AG2
 
 | | uPA-HMA | | Descriptor: | 3,5-bis(azanyl)-N-carbamimidoyl-6-(2-methoxypyrimidin-5-yl)pyrazine-2-carboxamide, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Buckley, B, Jiang, L.G, Huang, M.D, Kelso, M, Ranson, M. | | Deposit date: | 2018-08-09 | | Release date: | 2019-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | uPA-HMA
To Be Published
|
|
6AG7
 
 | | The crystal structure of uPA in complex with HMA-55F | | Descriptor: | 3,5-diamino-N-carbamimidoyl-6-(1-methyl-1H-pyrazol-4-yl)pyrazine-2-carboxamide, Urokinase-type plasminogen activator | | Authors: | Buckley, B, Jiang, L.G, Majed, H, Huang, M.D, Kelso, M, Ranson, M. | | Deposit date: | 2018-08-09 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | uPA-HMA
To Be Published
|
|
6ITE
 
 | | Crystal structure of group A Streptococcal surface dehydrogenase (SDH) | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Yuan, C, Li, R, Huang, M.D. | | Deposit date: | 2018-11-21 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | Structural determination of group A Streptococcal surface dehydrogenase and characterization of its interaction with urokinase-type plasminogen activator receptor.
Biochem.Biophys.Res.Commun., 510, 2019
|
|
8XDL
 
 | | F-actin-END | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Wang, L, Li, J, Liu, T, Huang, M. | | Deposit date: | 2023-12-11 | | Release date: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Aglycone polyethers target actin filaments as muscle-invasive bladder cancer inhibitors
To Be Published
|
|
8XDM
 
 | | F-actin-MAD | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Wang, L, Li, J, Liu, T, Huang, M. | | Deposit date: | 2023-12-11 | | Release date: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Aglycone polyethers target actin filaments as muscle-invasive bladder cancer inhibitors
To Be Published
|
|
5Z9Y
 
 | | Crystal structure of Mycobacterium tuberculosis thiazole synthase (ThiG) complexed with DXP | | Descriptor: | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE, Thiazole synthase | | Authors: | Zhang, J, Zhang, B, Zhao, Y, Yang, X, Huang, M, Cui, P, Zhang, W, Li, J, Zhang, Y. | | Deposit date: | 2018-02-05 | | Release date: | 2018-04-11 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Snapshots of catalysis: Structure of covalently bound substrate trapped in Mycobacterium tuberculosis thiazole synthase (ThiG).
Biochem. Biophys. Res. Commun., 497, 2018
|
|
5YC7
 
 | |
5Z1C
 
 | | The crystal structure of uPA in complex with 4-Iodobenzylamine at pH7.4 | | Descriptor: | 1-(4-iodophenyl)methanamine, Urokinase-type plasminogen activator | | Authors: | Jiang, L.G, Zhang, X, Luo, Z.P, Huang, M.D. | | Deposit date: | 2017-12-25 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Halogen bonding for the design of inhibitors by targeting the S1 pocket of serine proteases
Rsc Adv, 49, 2018
|
|
5ZLZ
 
 | | Structure of tPA and PAI-1 | | Descriptor: | GLYCEROL, Plasminogen activator inhibitor 1, Tissue-type plasminogen activator | | Authors: | Min, L, Huang, M. | | Deposit date: | 2018-03-31 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.581 Å) | | Cite: | Development of a PAI-1 trapping agent (PAItrap2) based on inactivated tPA-SPD and the crystal structure of PAItrap2 in complex with PAI-1
To Be Published
|
|
5YUP
 
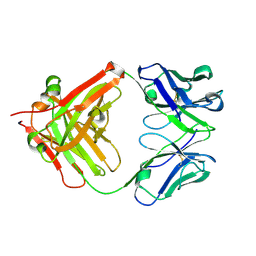 | | Crystal Structure of the Fab fragment of FVIIa antibody mAb4F5 | | Descriptor: | the heavy chain of the Fab fragment of FVIIa antibody mAb4F5, the light chain of the Fab fragment of FVIIa antibody mAb4F5 | | Authors: | Jiang, L.G, Persson, E, Huang, M.D. | | Deposit date: | 2017-11-22 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure, epitope, and functional impact of an antibody against a superactive FVIIa provide insights into allosteric mechanism.
Res Pract Thromb Haemost, 3, 2019
|
|
6KA4
 
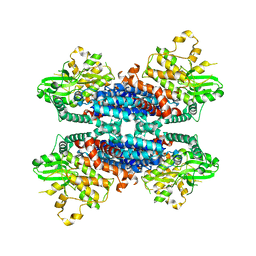 | | Cryo-EM structure of the AtMLKL3 tetramer | | Descriptor: | F22L4.1 protein | | Authors: | Lisa, M, Huang, M, Zhang, X, Ryohei, T.N, Leila, B.K, Isabel, M.L.S, Florence, J, Viera, K, Dmitry, L, Jane, E.P, James, M.M, Kay, H, Paul, S.L, Chai, J, Takaki, M. | | Deposit date: | 2019-06-20 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the AtMLKL3 tetramer
To Be Published
|
|
6LBA
 
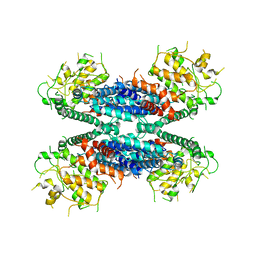 | | Cryo-EM structure of the AtMLKL2 tetramer | | Descriptor: | Protein kinase family protein | | Authors: | Lisa, M, Huang, M, Zhang, X, Ryohei, T.N, Leila, B.K, Isabel, M.L.S, Florence, J, Viera, K, Dmitry, L, Jane, E.P, James, M.M, Kay, H, Paul, S.L, Chai, J, Takaki, M. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the AtMLKL3 tetramer
To Be Published
|
|
7WOK
 
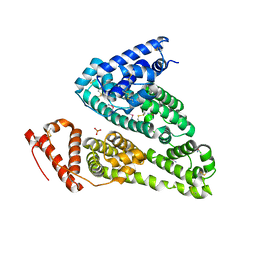 | | Crystal structure of HSA soaked with cisplatin for one week | | Descriptor: | Albumin, Cisplatin, PHOSPHATE ION | | Authors: | Chen, S.L, Yuan, C, Jiang, L.G, Luo, Z.P, Huang, M.D. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic analysis of interaction between cisplatin and human serum albumin: Effect of fatty acid.
Int.J.Biol.Macromol., 216, 2022
|
|
7WOJ
 
 | | Crystal structure of HSA-Myr complex soaked with cisplatin for one week | | Descriptor: | Albumin, Cisplatin, MYRISTIC ACID | | Authors: | Chen, S.L, Yuan, C, Jiang, L.G, Luo, Z.P, Huang, M.D. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystallographic analysis of interaction between cisplatin and human serum albumin: Effect of fatty acid.
Int.J.Biol.Macromol., 216, 2022
|
|
