8SRY
 
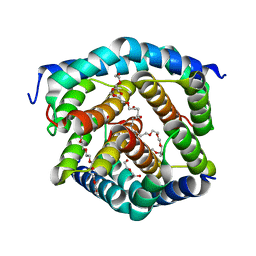 | | Crystal structure of BAK-BAX heterodimer with C12E8 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, ... | | Authors: | Brouwer, J.M, Czabotar, P.E, Colman, P.M, Miller, M.S. | | Deposit date: | 2023-05-08 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 2023
|
|
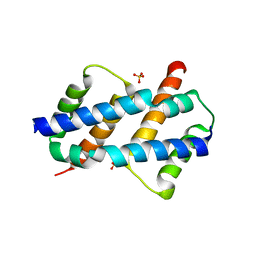
8SPZ
 
 | |
8SVK
 
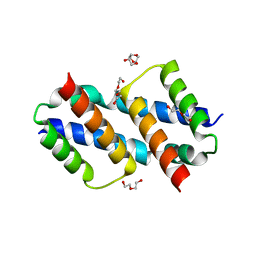 | |
6MCY
 
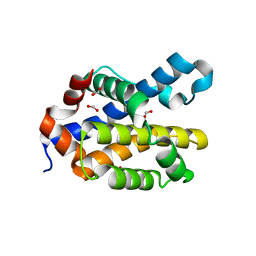 | | Crystal structure of mouse Bak | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2 homologous antagonist/killer, FORMIC ACID | | Authors: | Brouwer, J.M, Czabotar, P.E, Colman, P.M. | | Deposit date: | 2018-09-03 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | A small molecule interacts with VDAC2 to block mouse BAK-driven apoptosis.
Nat.Chem.Biol., 15, 2019
|
|
3QBR
 
 | | BakBH3 in complex with sjA | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Bcl-2 homologous antagonist/killer, SJCHGC06286 protein | | Authors: | Lee, E.F, Clarke, O.B, Fairlie, W.D, Colman, P.M, Evangelista, M, Feng, Z, Speed, T.P, Tchoubrieva, E, Strasser, A, Kalinna, B. | | Deposit date: | 2011-01-13 | | Release date: | 2011-04-13 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Discovery and molecular characterization of a Bcl-2-regulated cell death pathway in schistosomes.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3D7V
 
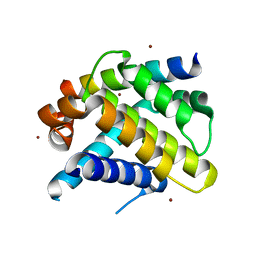 | | Crystal structure of Mcl-1 in complex with an Mcl-1 selective BH3 ligand | | Descriptor: | Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Lee, E.F, Czabotar, P.E, Colman, P.M, Fairlie, W.D. | | Deposit date: | 2008-05-22 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A novel BH3 ligand that selectively targets Mcl-1 reveals that apoptosis can proceed without Mcl-1 degradation.
J.Cell Biol., 180, 2008
|
|
3FDM
 
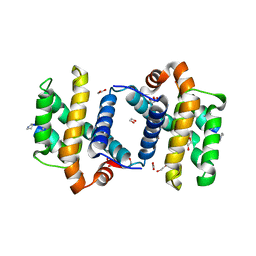 | | alpha/beta foldamer in complex with Bcl-xL | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator Bcl-X, alpha/beta-peptide foldamer | | Authors: | Fairlie, W.D, Lee, E.F, Smith, B.J, Czabotar, P.E, Colman, P.M, Sadowsky, J.D, Peterson-Kaufman, K.J, Gellman, S.H. | | Deposit date: | 2008-11-26 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | High-Resolution Structural Characterization of a Helical alpha/beta-Peptide Foldamer Bound to the Anti-Apoptotic Protein Bcl-x(L)
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
6O0L
 
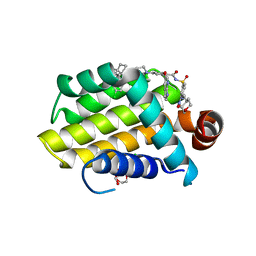 | | crystal structure of BCL-2 G101V mutation with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, CHLORIDE ION, ... | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-16 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
6O0P
 
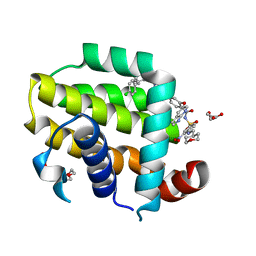 | | crystal structure of BCL-2 G101A mutation with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, DI(HYDROXYETHYL)ETHER | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-17 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
6O0O
 
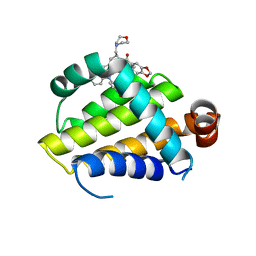 | | crystal structure of BCL-2 G101V mutation with S55746 | | Descriptor: | Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, ~{N}-(4-hydroxyphenyl)-3-[6-[[(3~{S})-3-(morpholin-4-ylmethyl)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]-1,3-benzodioxol-5-yl]-~{N}-phenyl-5,6,7,8-tetrahydroindolizine-1-carboxamide | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-17 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
6O0M
 
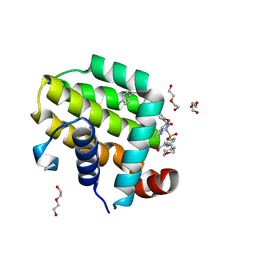 | | crystal structure of BCL-2 F104L mutation with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, DI(HYDROXYETHYL)ETHER | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-16 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
6O0K
 
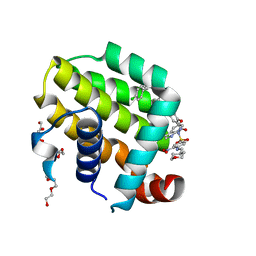 | | crystal structure of BCL-2 with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2, NONAETHYLENE GLYCOL | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-16 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
2BAT
 
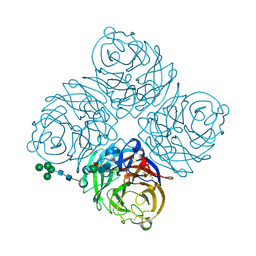 | | THE STRUCTURE OF THE COMPLEX BETWEEN INFLUENZA VIRUS NEURAMINIDASE AND SIALIC ACID, THE VIRAL RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-08-10 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the complex between influenza virus neuraminidase and sialic acid, the viral receptor.
Proteins, 14, 1992
|
|
4C52
 
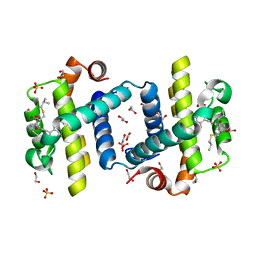 | | Crystal structure of Bcl-xL in complex with benzoylurea compound (39b) | | Descriptor: | (R)-2-(3-(3-((2,4-DIFLUOROPENYL)ETHYNYL)BENZOYL)-3-PROPYLUREIDO)-3-(ISOBUTYLTHIO) PROPANOIC ACID, 1,2-ETHANEDIOL, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Roy, M.J, Brady, R.M, Lessene, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2013-09-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | De-Novo Designed Library of Benzoylureas as Inhibitors of Bcl-Xl: Synthesis, Structural and Biochemical Characterization.
J.Med.Chem., 57, 2014
|
|
4C5D
 
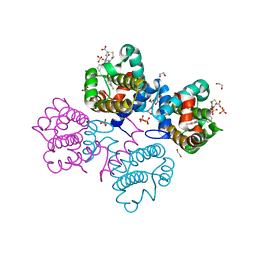 | | Crystal structure of Bcl-xL in complex with benzoylurea compound (42) | | Descriptor: | (R)-3-(4-BROMOBENZYLTHIO)-2-(3-(3-((2,4-DIFLUOROPHENYL)ETHYNYL)BENZOYL)-3-PROPYLUREIDO)PROPANOIC ACID, 1,2-ETHANEDIOL, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Roy, M.J, Brady, R.M, Lessene, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | De-Novo Designed Library of Benzoylureas as Inhibitors of Bcl-Xl: Synthesis, Structural and Biochemical Characterization.
J.Med.Chem., 57, 2014
|
|
2XPX
 
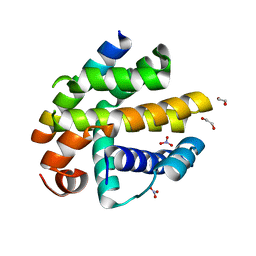 | | Crystal structure of BHRF1:Bak BH3 complex | | Descriptor: | 1,2-ETHANEDIOL, APOPTOSIS REGULATOR BHRF1, BCL-2 HOMOLOGOUS ANTAGONIST/KILLER, ... | | Authors: | Kvansakul, M, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2010-08-31 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for Apoptosis Inhibition by Epstein-Barr Virus Bhrf1.
Plos Pathog., 6, 2010
|
|
2Y6W
 
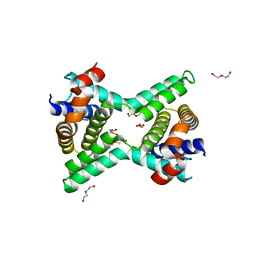 | | Structure of a Bcl-w dimer | | Descriptor: | BCL-2-LIKE PROTEIN 2, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | Authors: | Lee, E.F, Evangelista, M, Pettikiriarachchi, A, Dogovski, C, Perugini, M.A, Colman, P.M, Fairlie, W.D. | | Deposit date: | 2011-01-27 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Bcl-W Domain-Swapped Dimer: Implications for the Function of Bcl-2 Family Proteins.
Structure, 19, 2011
|
|
2YJ1
 
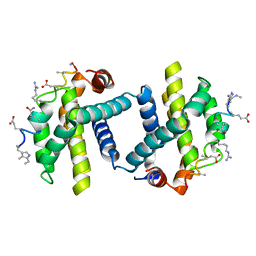 | | Puma BH3 foldamer in complex with Bcl-xL | | Descriptor: | ALPHA-BETA-PUMA BH3 FOLDAMER, BCL-2-LIKE PROTEIN 1 | | Authors: | Lee, E.F, Smith, B.J, Horne, W.S, Mayer, K.N, Evangelista, M, Colman, P.M, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2011-05-18 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Basis of Bcl-Xl Recognition by a Bh3-Mimetic Alpha-Beta-Peptide Generated Via Sequence-Based Design
Chembiochem, 12, 2011
|
|
3FDL
 
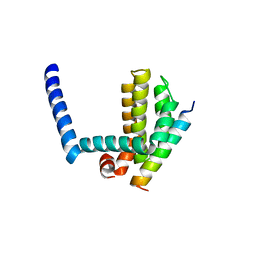 | | Bim BH3 peptide in complex with Bcl-xL | | Descriptor: | Apoptosis regulator Bcl-X, Bcl-2-like protein 11 | | Authors: | Fairlie, W.D, Lee, E.F, Smith, B.J, Czabotar, P.E, Colman, P.M. | | Deposit date: | 2008-11-26 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | High-Resolution Structural Characterization of a Helical alpha/beta-Peptide Foldamer Bound to the Anti-Apoptotic Protein Bcl-x(L)
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
4ZIE
 
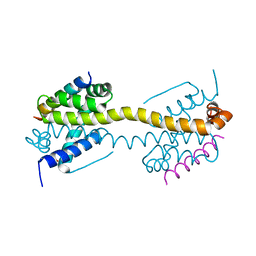 | | Crystal Structure of core/latch dimer of Bax in complex with BimBH3 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX, Bcl-2-like protein 11 | | Authors: | Krishna Kumar, K, Robin, A.Y, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
4ZIH
 
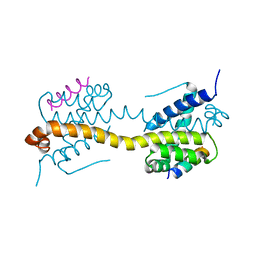 | | Crystal Structure of core/latch dimer of Bax in complex with BimBH3mini | | Descriptor: | Apoptosis regulator BAX, Bcl-2-like protein 11 | | Authors: | Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
4ZIF
 
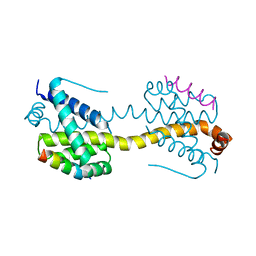 | | Crystal Structure of core/latch dimer of Bax in complex with BimBH3mini | | Descriptor: | Apoptosis regulator BAX, Bcl-2-like protein 11 | | Authors: | Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
4ZII
 
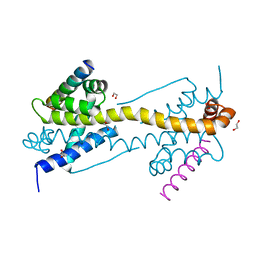 | | Crystal Structure of core/latch dimer of BaxI66A in complex with BidBH3 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX, BH3-interacting domain death agonist | | Authors: | Czabotar, P.E, Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
4ZIG
 
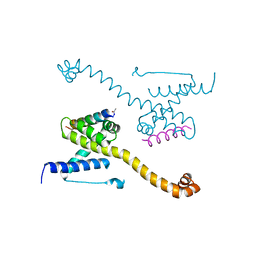 | | Crystal Structure of core/latch dimer of Bax in complex with BidBH3mini | | Descriptor: | Apoptosis regulator BAX, BH3-interacting domain death agonist | | Authors: | Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
6UXM
 
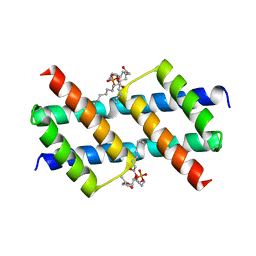 | |
