3PS1
 
 | | Crystal structure of the Escherichia Coli LPXC/LPC-011 complex | | 分子名称: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-1-(hydroxyamino)-1-oxobutan-2-yl]benzamide, 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2010-11-30 | | 公開日 | 2011-01-19 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Syntheses, structures and antibiotic activities of LpxC inhibitors based on the diacetylene scaffold.
Bioorg.Med.Chem., 19, 2011
|
|
3PS2
 
 | | Crystal structure of the Escherichia Coli LPXC/LPC-012 complex | | 分子名称: | 4-[4-(3-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-1-nitroso-1-oxobutan-2-yl]benzamide, 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2010-11-30 | | 公開日 | 2011-01-19 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Syntheses, structures and antibiotic activities of LpxC inhibitors based on the diacetylene scaffold.
Bioorg.Med.Chem., 19, 2011
|
|
3PS3
 
 | | Crystal structure of the Escherichia Coli LPXC/LPC-053 complex | | 分子名称: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3S)-3-hydroxy-1-nitroso-1-oxobutan-2-yl]benzamide, 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2010-11-30 | | 公開日 | 2011-01-19 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Syntheses, structures and antibiotic activities of LpxC inhibitors based on the diacetylene scaffold.
Bioorg.Med.Chem., 19, 2011
|
|
3QC1
 
 | | Protein Phosphatase Subunit: Alpha4 | | 分子名称: | Immunoglobulin-binding protein 1 | | 著者 | Spiller, B.W, LeNoue-Newton, M.L, Watkins, G.R, Germane, K.L, Zhou, P, McCorvey, L.R, Wadzinski, B.E. | | 登録日 | 2011-01-14 | | 公開日 | 2011-03-30 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | The Mid1 and PP2Ac binding domains of Alpha4 are both required for Alpha4 to inhibit PP2Ac degradation
TO BE PUBLISHED
|
|
7CAL
 
 | | Cryo-EM Structure of the Hyperpolarization-Activated Inwardly Rectifying Potassium Channel KAT1 from Arabidopsis | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Potassium channel KAT1 | | 著者 | Li, S.Y, Yang, F, Sun, D.M, Zhang, Y, Zhang, M.G, Zhou, P, Liu, S.L, Zhang, Y.N, Zhang, L.H, Tian, C.L. | | 登録日 | 2020-06-09 | | 公開日 | 2020-07-29 | | 最終更新日 | 2020-11-18 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structure of the hyperpolarization-activated inwardly rectifying potassium channel KAT1 from Arabidopsis.
Cell Res., 30, 2020
|
|
4MQY
 
 | | Crystal Structure of the Escherichia coli LpxC/LPC-138 complex | | 分子名称: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-2-methyl-1-nitroso-1-oxobutan-2-yl]benzamide, 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, ... | | 著者 | Lee, C.-J, Najeeb, J, Zhou, P. | | 登録日 | 2013-09-17 | | 公開日 | 2013-10-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.005 Å) | | 主引用文献 | Structural Basis of the Promiscuous Inhibitor Susceptibility of Escherichia coli LpxC.
Acs Chem.Biol., 9, 2014
|
|
4ISA
 
 | | Crystal Structure of the Escherichia coli LpxC/BB-78485 complex | | 分子名称: | (2R)-N-hydroxy-3-naphthalen-2-yl-2-[(naphthalen-2-ylsulfonyl)amino]propanamide, (4S,5S)-1,2-DITHIANE-4,5-DIOL, FORMIC ACID, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2013-01-16 | | 公開日 | 2013-10-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Basis of the Promiscuous Inhibitor Susceptibility of Escherichia coli LpxC.
Acs Chem.Biol., 9, 2014
|
|
4IS9
 
 | | Crystal Structure of the Escherichia coli LpxC/L-161,240 complex | | 分子名称: | (4R)-2-(3,4-dimethoxy-5-propylphenyl)-N-hydroxy-4,5-dihydro-1,3-oxazole-4-carboxamide, ISOPROPYL ALCOHOL, SODIUM ION, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2013-01-16 | | 公開日 | 2013-10-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Structural Basis of the Promiscuous Inhibitor Susceptibility of Escherichia coli LpxC.
Acs Chem.Biol., 9, 2014
|
|
4ITL
 
 | | Crystal structure of LpxK from Aquifex aeolicus in complex with AMP-PCP at 2.1 angstrom resolution | | 分子名称: | GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Tetraacyldisaccharide 4'-kinase | | 著者 | Emptage, R.P, Pemble IV, C.W, York, J.D, Raetz, C.R.H, Zhou, P. | | 登録日 | 2013-01-18 | | 公開日 | 2013-04-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Mechanistic Characterization of the Tetraacyldisaccharide-1-phosphate 4'-Kinase LpxK Involved in Lipid A Biosynthesis.
Biochemistry, 52, 2013
|
|
4ITN
 
 | | Crystal structure of "compact P-loop" LpxK from Aquifex aeolicus in complex with chloride at 2.2 angstrom resolution | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | 著者 | Emptage, R.P, Pemble IV, C.W, York, J.D, Raetz, C.R.H, Zhou, P. | | 登録日 | 2013-01-18 | | 公開日 | 2013-04-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1912 Å) | | 主引用文献 | Mechanistic Characterization of the Tetraacyldisaccharide-1-phosphate 4'-Kinase LpxK Involved in Lipid A Biosynthesis.
Biochemistry, 52, 2013
|
|
4ITM
 
 | | Crystal structure of "apo" form LpxK from Aquifex aeolicus in complex with ATP at 2.2 angstrom resolution | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Emptage, R.P, Pemble IV, C.W, York, J.D, Raetz, C.R.H, Zhou, P. | | 登録日 | 2013-01-18 | | 公開日 | 2013-04-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1994 Å) | | 主引用文献 | Mechanistic Characterization of the Tetraacyldisaccharide-1-phosphate 4'-Kinase LpxK Involved in Lipid A Biosynthesis.
Biochemistry, 52, 2013
|
|
4J72
 
 | | Crystal Structure of polyprenyl-phosphate N-acetyl hexosamine 1-phosphate transferase | | 分子名称: | MAGNESIUM ION, NICKEL (II) ION, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | 著者 | Lee, S.Y, Chung, B.C, Gillespie, R.A, Kwon, D.Y, Guan, Z, Zhou, P, Hong, J. | | 登録日 | 2013-02-12 | | 公開日 | 2013-09-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis.
Science, 341, 2013
|
|
4LCG
 
 | | Crystal structure of the Pseudomonas aeruginosa LPXC/LPC-050 complex | | 分子名称: | (betaS)-Nalpha-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N,beta-dihydroxy-L-tyrosinamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2013-06-21 | | 公開日 | 2013-08-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.568 Å) | | 主引用文献 | Synthesis, Structure, and Antibiotic Activity of Aryl-Substituted LpxC Inhibitors.
J.Med.Chem., 56, 2013
|
|
4LCH
 
 | | Crystal structure of the Pseudomonas aeruginosa LPXC/LPC-051 complex | | 分子名称: | (betaS)-Nalpha-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N,beta-dihydroxy-beta-methyl-L-tyrosinamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2013-06-21 | | 公開日 | 2013-08-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.596 Å) | | 主引用文献 | Synthesis, Structure, and Antibiotic Activity of Aryl-Substituted LpxC Inhibitors.
J.Med.Chem., 56, 2013
|
|
4LCF
 
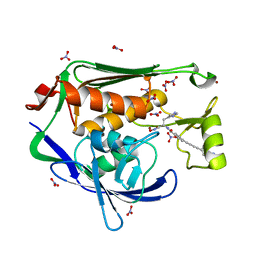 | | Crystal structure of the Pseudomonas aeruginosa LPXC/LPC-014 complex | | 分子名称: | NITRATE ION, Nalpha-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N-hydroxy-L-histidinamide, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2013-06-21 | | 公開日 | 2013-08-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.599 Å) | | 主引用文献 | Synthesis, Structure, and Antibiotic Activity of Aryl-Substituted LpxC Inhibitors.
J.Med.Chem., 56, 2013
|
|
5DRO
 
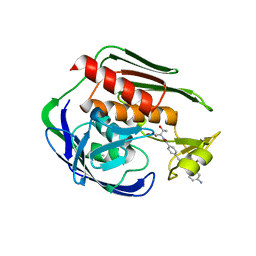 | | Structure of the Aquifex aeolicus LpxC/LPC-011 Complex | | 分子名称: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-1-(hydroxyamino)-1-oxobutan-2-yl]benzamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Lee, C.-J, Najeeb, J, Zhou, P. | | 登録日 | 2015-09-16 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
5DRP
 
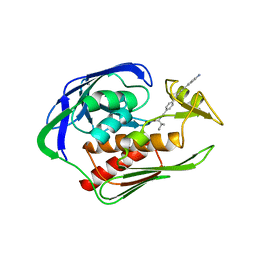 | | Structure of the AaLpxC/LPC-023 Complex | | 分子名称: | CHLORIDE ION, DIMETHYL SULFOXIDE, N~2~-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N-hydroxy-L-isoleucinamide, ... | | 著者 | Najeeb, J, Lee, C.-J, Zhou, P. | | 登録日 | 2015-09-16 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.889 Å) | | 主引用文献 | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
4LKV
 
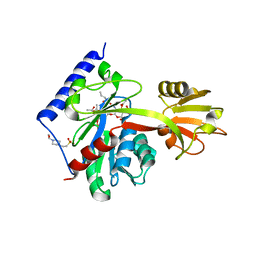 | | Determinants of lipid substrate and membrane binding for the tetraacyldisaccharide-1-phosphate 4 -kinase LpxK | | 分子名称: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-deoxy-3-O-[(3R)-3-hydroxytetradecanoyl]-2-{[(3R)-3-hydroxytetradecanoyl]amino}-4-O-phosphono-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Emptage, R.P, Tonthat, N.K, York, J.D, Schumacher, M.A, Zhou, P. | | 登録日 | 2013-07-08 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.5109 Å) | | 主引用文献 | Structural Basis of Lipid Binding for the Membrane-embedded Tetraacyldisaccharide-1-phosphate 4'-Kinase LpxK.
J.Biol.Chem., 289, 2014
|
|
5DRR
 
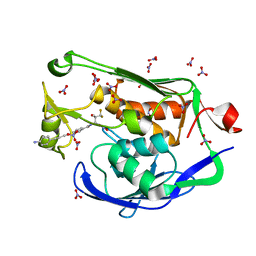 | | Crystal structure of the Pseudomonas aeruginosa LpxC/LPC-058 complex | | 分子名称: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3S)-4,4-difluoro-3-hydroxy-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]benzamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | 著者 | Lee, C.-J, Najeeb, J, Zhou, P. | | 登録日 | 2015-09-16 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
5DRQ
 
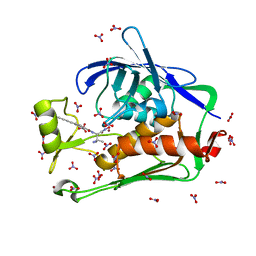 | | Crystal structure of the Pseudomonas aeruginosa LpxC/LPC-040 complex | | 分子名称: | N-[(2S)-3-amino-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]-4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | 著者 | Lee, C.-J, Najeeb, J, Zhou, P. | | 登録日 | 2015-09-16 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
2J4I
 
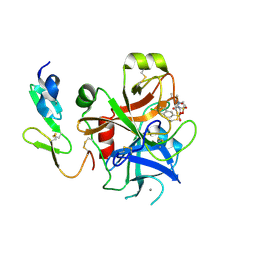 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | 分子名称: | 1-PYRROLIDINEACETAMIDE, 3-[[(6-CHLORO-2-NAPHTHALENYL)SULFONYL]AMINO]-ALPHA-METHYL-N-(1-METHYLETHYL)-N-[2-[(METHYLSULFONYL)AMINO]ETHYL]-2-OXO-, (ALPHAS,3S)-, ... | | 著者 | Young, R.J, Campbell, M, Borthwick, A.D, Brown, D, Chan, C, Convery, M.A, Crowe, M.C, Dayal, S, Diallo, H, Kelly, H.A, Paul King, N, Kleanthous, S, Kurtis, C.L, Mason, A.M, Mordaunt, J.E, Patel, C, Pateman, A.J, Senger, S, Shah, G.P, Smith, P.W, Watson, N.S, Weston, H.E, Zhou, P. | | 登録日 | 2006-08-31 | | 公開日 | 2006-09-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure- and Property-Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Acyclic Alanyl Amides as P4 Motifs.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2J95
 
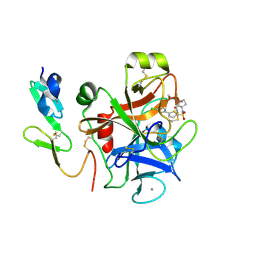 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | 分子名称: | 5'-CHLORO-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}-2,2'-BITHIOPHENE-5-SULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, ACTIVATED FACTOR XA LIGHT CHAIN, ... | | 著者 | Chan, C, Borthwick, A.D, Brown, D, Campbell, M, Chaudry, L, Chung, C.W, Convery, M.A, Hamblin, J.N, Johnstone, L, Kelly, H.A, Kleanthous, S, Burns-Kurtis, C.L, Patikis, A, Patel, C, Pateman, A.J, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E, Whitworth, C, Young, R.J, Zhou, P. | | 登録日 | 2006-11-02 | | 公開日 | 2007-03-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Factor Xa Inhibitors: S1 Binding Interactions of a Series of N-{(3S)-1-[(1S)-1-Methyl-2-Morpholin-4-Yl-2-Oxoethyl]-2-Oxopyrrolidin-3-Yl}Sulfonamides.
J.Med.Chem., 50, 2007
|
|
2J94
 
 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | 分子名称: | 5-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}-1H-1,2,4-TRIAZOLE-3-SULFONAMIDE, CALCIUM ION, COAGULATION FACTOR X | | 著者 | Chan, C, Borthwick, A.D, Brown, D, Campbell, M, Chaudry, L, Chung, C.W, Convery, M.A, Hamblin, J.N, Johnstone, L, Kelly, H.A, Kleanthous, S, Burns-Kurtis, C.L, Patikis, A, Patel, C, Pateman, A.J, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E, Whitworth, C, Young, R.J, Zhou, P. | | 登録日 | 2006-11-02 | | 公開日 | 2007-03-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Factor Xa Inhibitors: S1 Binding Interactions of a Series of N-{(3S)-1-[(1S)-1-Methyl-2-Morpholin-4-Yl-2-Oxoethyl]-2-Oxopyrrolidin-3-Yl}Sulfonamides.
J.Med.Chem., 50, 2007
|
|
2JT2
 
 | | Solution Structure of the Aquifex aeolicus LpxC- CHIR-090 complex | | 分子名称: | N-{(1S,2R)-2-hydroxy-1-[(hydroxyamino)carbonyl]propyl}-4-{[4-(morpholin-4-ylmethyl)phenyl]ethynyl}benzamide, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | 著者 | Barb, A.W, Jiang, L, Raetz, C.R.H, Zhou, P. | | 登録日 | 2007-07-18 | | 公開日 | 2007-12-04 | | 最終更新日 | 2021-10-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the deacetylase LpxC bound to the antibiotic CHIR-090: Time-dependent inhibition and specificity in ligand binding
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2LSG
 
 | |
