4TXA
 
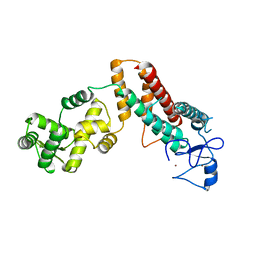 | |
4XBM
 
 | | X-ray crystal structure of Notch ligand Delta-like 1 | | Descriptor: | Delta-like protein 1, alpha-L-fucopyranose | | Authors: | Kershaw, N.J, Burgess, A.W, Church, N.L, Luo, C.S, Adam, T.E. | | Deposit date: | 2014-12-17 | | Release date: | 2015-03-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Notch ligand delta-like1: X-ray crystal structure and binding affinity.
Biochem.J., 468, 2015
|
|
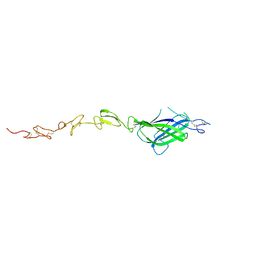
4GL9
 
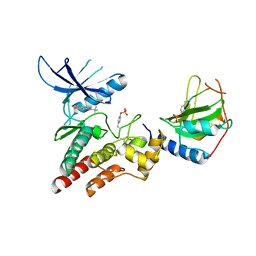 | | Crystal structure of inhibitory protein SOCS3 in complex with JAK2 kinase domain and fragment of GP130 intracellular domain | | Descriptor: | 2-TERT-BUTYL-9-FLUORO-3,6-DIHYDRO-7H-BENZ[H]-IMIDAZ[4,5-F]ISOQUINOLINE-7-ONE, Interleukin-6 receptor subunit beta, PHOSPHATE ION, ... | | Authors: | Kershaw, N.J, Murphy, J.M, Laktyushin, A, Nicola, N.A, Babon, J.J. | | Deposit date: | 2012-08-14 | | Release date: | 2013-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | SOCS3 binds specific receptor-JAK complexes to control cytokine signaling by direct kinase inhibition.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6BZH
 
 | | Structure of mouse RIG-I tandem CARDs | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Probable ATP-dependent RNA helicase DDX58 | | Authors: | Kershaw, N.J, D'Cruz, A.A, Babon, J.J, Nicholson, S.E. | | Deposit date: | 2017-12-24 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a second binding site on the TRIM25 B30.2 domain.
Biochem. J., 475, 2018
|
|
3L5I
 
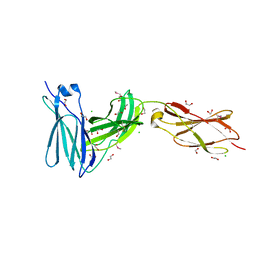 | | Crystal structure of FnIII domains of human GP130 (Domains 4-6) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Interleukin-6 receptor subunit beta | | Authors: | Kershaw, N.J, Zhang, J.-G, Garrett, T.P.J, Czabotar, P.E. | | Deposit date: | 2009-12-22 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the entire ectodomain of gp130: insights into the molecular assembly of the tall cytokine receptor complexes.
J.Biol.Chem., 285, 2010
|
|
3L5J
 
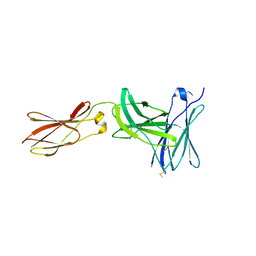 | | Crystal structure of FnIII domains of human GP130 (Domains 4-6) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Interleukin-6 receptor subunit beta | | Authors: | Kershaw, N.J, Zhang, J.-G, Garrett, T.P.J, Czabotar, P.E. | | Deposit date: | 2009-12-22 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.042 Å) | | Cite: | Crystal structure of the entire ectodomain of gp130: insights into the molecular assembly of the tall cytokine receptor complexes.
J.Biol.Chem., 285, 2010
|
|
7M6T
 
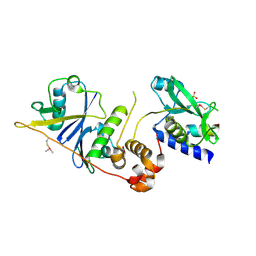 | | Crystal structure of SOCS2/ElonginB/ElonginC bound to a non-canonical peptide that enhances phospho-peptide binding | | Descriptor: | Elongin-B, Elongin-C, Non-canonical peptide F3, ... | | Authors: | Kershaw, N.J, Li, K, Linossi, E.M, Nicholson, S.E. | | Deposit date: | 2021-03-26 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Discovery of an exosite on the SOCS2-SH2 domain that enhances SH2 binding to phosphorylated ligands.
Nat Commun, 12, 2021
|
|
8SKL
 
 | | PTP1B in complex with 182 | | Descriptor: | 1,2-ETHANEDIOL, 5-[1-fluoro-3-hydroxy-7-(3-hydroxy-3-methylbutoxy)naphthalen-2-yl]-1lambda~6~,2,5-thiadiazolidine-1,1,3-trione, CHLORIDE ION, ... | | Authors: | Kershaw, N.J, Babon, J.J, Chen, H, Tiganis, T. | | Deposit date: | 2023-04-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A small molecule inhibitor of PTP1B and PTPN2 enhances T cell anti-tumor immunity.
Nat Commun, 14, 2023
|
|
6C5X
 
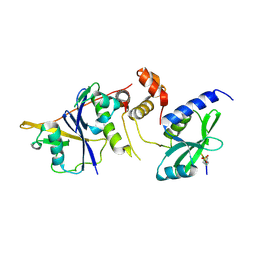 | | Crystal Structure of SOCS1 in complex with ElonginB and ElonginC | | Descriptor: | Elongin-B, Elongin-C, GP130 peptide fragment, ... | | Authors: | Kershaw, N.J, Laktyushin, A, Babon, J.J. | | Deposit date: | 2018-01-17 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.105 Å) | | Cite: | The molecular basis of JAK/STAT inhibition by SOCS1.
Nat Commun, 9, 2018
|
|
4B8E
 
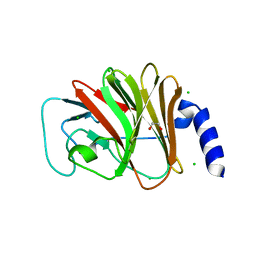 | | PRY-SPRY domain of Trim25 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, E3 UBIQUITIN/ISG15 LIGASE TRIM25 | | Authors: | Kershaw, N.J, D'Cruz, A.A, Nicola, N.A, Nicholson, S.E, Babon, J.J. | | Deposit date: | 2012-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Crystal Structure of the Trim25 B30.2 (Pryspry) Domain: A Key Component of Antiviral Signaling.
Biochem.J., 456, 2013
|
|
1VZ6
 
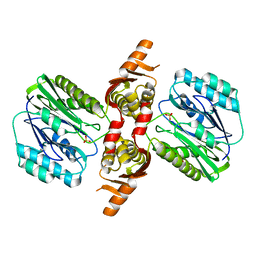 | |
7R8W
 
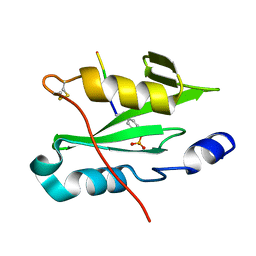 | |
7R8X
 
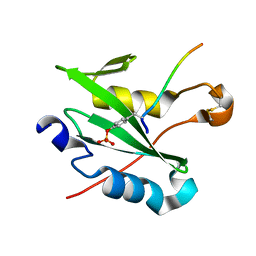 | |
8CZ9
 
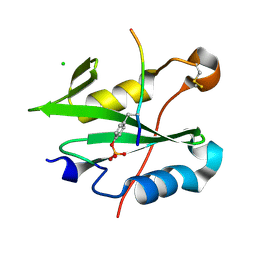 | |
2XFT
 
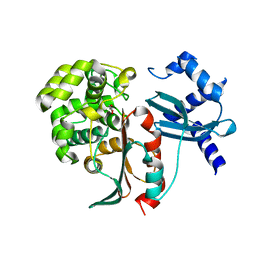 | | Structural and mechanistic studies on a cephalosporin esterase from the clavulanic acid biosynthesis pathway | | Descriptor: | GLYCEROL, ORF12 | | Authors: | Iqbal, A, Valegard, K, Kershaw, N.J, Ivison, D, Genereux, C, Dubus, A, Andersson, I, Schofield, C.J. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
2XFS
 
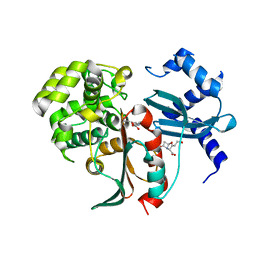 | | Structural and mechanistic studies on a cephalosporin esterase from the clavulanic acid biosynthesis pathway | | Descriptor: | (2R,3Z,5R)-3-(2-HYDROXYETHYLIDENE)-7-OXO-4-OXA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, ORF12 | | Authors: | Iqbal, A, Valegard, K, Kershaw, N.J, Ivison, D, Genereux, C, Dubus, A, Andersson, I, Schofield, C.J. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
8U18
 
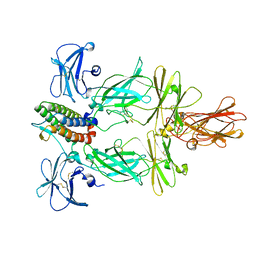 | | Cryo-EM structure of murine Thrombopoietin receptor ectodomain in complex with Tpo | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Thrombopoietin, Thrombopoietin receptor,GCN4 isoform 1, ... | | Authors: | Sarson-Lawrence, K.S, Hardy, J.M, Leis, A, Babon, J.J, Kershaw, N.J. | | Deposit date: | 2023-08-30 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the extracellular domain of murine Thrombopoietin Receptor in complex with Thrombopoietin.
Nat Commun, 15, 2024
|
|
1VZ7
 
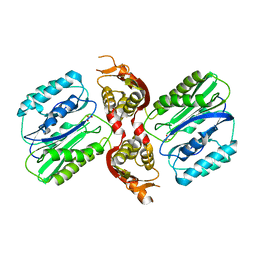 | |
1VZ8
 
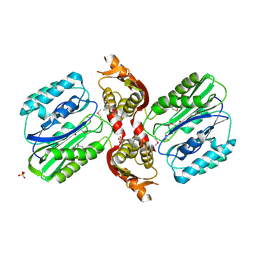 | |
2JAH
 
 | | Biochemical and structural analysis of the Clavulanic acid dehydeogenase (CAD) from Streptomyces clavuligerus | | Descriptor: | CLAVULANIC ACID DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | MacKenzie, A.K, Kershaw, N.J, Hernandez, H, Robinson, C.V, Schofield, C.J, Andersson, I. | | Deposit date: | 2006-11-28 | | Release date: | 2007-02-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Clavulanic Acid Dehydrogenase: Structural and Biochemical Analysis of the Final Step in the Biosynthesis of the Beta-Lactamase Inhibitor Clavulanic Acid
Biochemistry, 46, 2007
|
|
2JAP
 
 | | Clavulanic Acid Dehydrogenase: Structural and Biochemical Analysis of the Final Step in the Biosynthesis of the beta-Lactamase Inhibitor Clavulanic acid | | Descriptor: | (2R,3Z,5R)-3-(2-HYDROXYETHYLIDENE)-7-OXO-4-OXA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, CLAVALDEHYDE DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | MacKenzie, A.K, Kershaw, N.J, Hernandez, H, Robinson, C.V, Schofield, C.J, Andersson, I. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Clavulanic Acid Dehydrogenase: Structural and Biochemical Analysis of the Final Step in the Biosynthesis of the Beta-Lactamase Inhibitor Clavulanic Acid
Biochemistry, 46, 2007
|
|
6C7Y
 
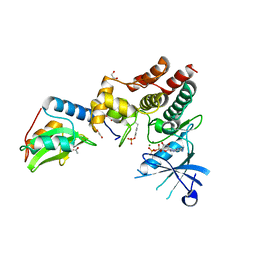 | | Crystal structure of inhibitory protein SOCS1 in complex with JAK1 kinase domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Liau, N.P.D, Laktyushin, A, Lucet, I.S, Murphy, J.M, Yao, S, Callaghan, K, Nicola, N.A, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2018-01-23 | | Release date: | 2018-05-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | The molecular basis of JAK/STAT inhibition by SOCS1.
Nat Commun, 9, 2018
|
|
8EXI
 
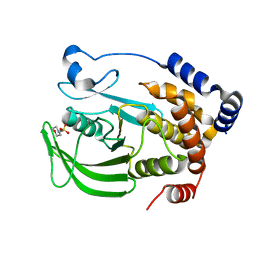 | |
8EXJ
 
 | | Crystal structure of PTP1B D181A/Q262A phosphatase domain in complex with a JAK1 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, Tyrosine-protein kinase JAK1 activation loop peptide, ... | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
8EXN
 
 | | Crystal structure of PTP1B D181A/Q262A phosphatase domain with TYK2 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Non-receptor tyrosine-protein kinase TYK2 activation loop peptide, PHOSPHATE ION, ... | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
