4UMY
 
 | | IDH1 R132H in complex with cpd 1 | | 分子名称: | GLYCEROL, ISOCITRATE DEHYDROGENASE [NADP] CYTOPLASMIC, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | McLean, L, Zhang, Y, Mathieu, M. | | 登録日 | 2014-05-22 | | 公開日 | 2014-11-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Selective Inhibition of Mutant Isocitrate Dehydrogenase 1 (Idh1) Via Disruption of a Metal Binding Network by an Allosteric Small Molecule.
J.Biol.Chem., 290, 2015
|
|
7SFR
 
 | | Unmethylated Mtb Ribosome 50S with SEQ-9 | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Xing, Z, Cui, Z, Zhang, J, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2021-10-04 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Discovery of natural-product-derived sequanamycins as potent oral anti-tuberculosis agents.
Cell, 186, 2023
|
|
6RO0
 
 | |
7KGB
 
 | |
4F0I
 
 | | Crystal structure of apo TrkA | | 分子名称: | High affinity nerve growth factor receptor | | 著者 | Liu, J. | | 登録日 | 2012-05-04 | | 公開日 | 2012-09-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.302 Å) | | 主引用文献 | The Crystal Structures of TrkA and TrkB Suggest Key Regions for Achieving Selective Inhibition.
J.Mol.Biol., 423, 2012
|
|
6I9J
 
 | | Human transforming growth factor beta2 in a tetragonal crystal form | | 分子名称: | Transforming growth factor beta-2 proprotein | | 著者 | Gomis-Ruth, F.X, Marino-Puertas, L, del Amo-Maestro, L, Goulas, T. | | 登録日 | 2018-11-23 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Recombinant production, purification, crystallization, and structure analysis of human transforming growth factor beta 2 in a new conformation.
Sci Rep, 9, 2019
|
|
7ZVT
 
 | | CryoEM structure of Ku heterodimer bound to DNA | | 分子名称: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*TP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | 登録日 | 2022-05-17 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
7Z6O
 
 | | X-Ray studies of Ku70/80 reveal the binding site for IP6 | | 分子名称: | DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*AP*AP*AP*CP*TP*AP*AP*AP*AP*AP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Varela, P.F, Charbonnier, J.B. | | 登録日 | 2022-03-14 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
7ZT6
 
 | |
5LO5
 
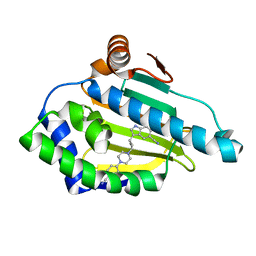 | | HSP90 WITH indole derivative | | 分子名称: | 3-[4-[4-(4-cyanophenyl)piperazin-1-yl]butyl]-6-oxidanyl-1~{H}-indole-5-carbonitrile, Heat shock protein HSP 90-alpha | | 著者 | Graedler, U, Amaral, M. | | 登録日 | 2016-08-08 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
5LO6
 
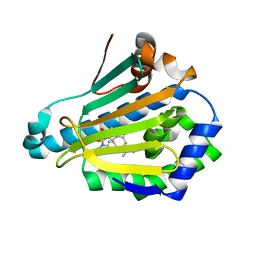 | | HSP90 WITH indazole derivative | | 分子名称: | 3-(3,3-dimethylbutyl)-~{N}-methyl-~{N}-[4-(1-methylpiperidin-4-yl)phenyl]-6-oxidanyl-2~{H}-indazole-5-carboxamide, Heat shock protein HSP 90-alpha | | 著者 | Graedler, U, Amaral, M. | | 登録日 | 2016-08-08 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
5HNI
 
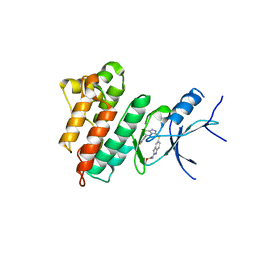 | | CRYSTAL STRUCTURE OF CMET WT with compound 3 | | 分子名称: | Hepatocyte growth factor receptor, methyl (6-{[6-(4-fluorophenyl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1H-benzimidazol-2-yl)carbamate | | 著者 | Vallee, F, Houtmann, J. | | 登録日 | 2016-01-18 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
5HO6
 
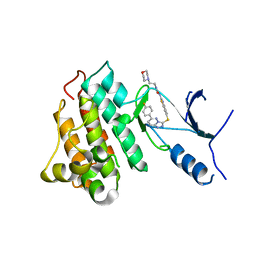 | | CRYSTAL STRUCTURE OF CMET IN COMPLEX WITH CMPD. | | 分子名称: | 1-(6-{[6-(4-fluorophenyl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1,3-benzothiazol-2-yl)-3-[2-(morpholin-4-yl)ethyl]urea, Hepatocyte growth factor receptor | | 著者 | Vallee, F, Houtmann, J. | | 登録日 | 2016-01-19 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
5HOR
 
 | | Crystal structure of c-Met-M1250T in complex with SAR125844. | | 分子名称: | 1-(6-{[6-(4-fluorophenyl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1,3-benzothiazol-2-yl)-3-[2-(morpholin-4-yl)ethyl]urea, Hepatocyte growth factor receptor | | 著者 | Vallee, F, Houtmann, J, Marquette, J.-P. | | 登録日 | 2016-01-19 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
5HLW
 
 | | Crystal structure of c-Met mutant Y1230H in complex with compound 14 | | 分子名称: | 1-[2-(1-ethylpiperidin-4-yl)ethyl]-3-(6-{[6-(thiophen-2-yl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1,3-benzothiazol-2-yl)urea, CHLORIDE ION, Hepatocyte growth factor receptor | | 著者 | Vallee, F, Pouzieux, S, Marquette, J.P, Houtmann, J. | | 登録日 | 2016-01-15 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
5HOA
 
 | | Crystal structure of c-Met L1195V in complex with SAR125844 | | 分子名称: | 1-(6-{[6-(4-fluorophenyl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1,3-benzothiazol-2-yl)-3-[2-(morpholin-4-yl)ethyl]urea, Hepatocyte growth factor receptor | | 著者 | Vallee, F, Marquette, J.-P. | | 登録日 | 2016-01-19 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
4KV5
 
 | | scFv GC1009 in complex with TGF-beta1. | | 分子名称: | Single-chain variable fragment GC1009, Transforming growth factor beta-1 proprotein | | 著者 | Wei, R, Moulin, A.G, Mathieu, M. | | 登録日 | 2013-05-22 | | 公開日 | 2014-09-24 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structures of a pan-specific antagonist antibody complexed to different isoforms of TGF beta reveal structural plasticity of antibody-antigen interactions.
Protein Sci., 23, 2014
|
|
4KXZ
 
 | | crystal structure of tgfb2 in complex with GC2008. | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, GC1008 Heavy Chain, ... | | 著者 | Mathieu, M, Moulin, A.G, Wei, R. | | 登録日 | 2013-05-28 | | 公開日 | 2014-09-24 | | 最終更新日 | 2014-12-03 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Structures of a pan-specific antagonist antibody complexed to different isoforms of TGF beta reveal structural plasticity of antibody-antigen interactions.
Protein Sci., 23, 2014
|
|
7AZO
 
 | | 70S thermus thermophilus ribosome with bound antibiotic lead SEQ-977 | | 分子名称: | (2R,3S,4R,5R,7S,9S,10S,11R,12S,13R)-12-(((2R,4R,5S,6S)-4,5-dihydroxy-4,6-dimethyltetrahydro-2H-pyran-2-yl)oxy)-2-((S)-1-(((2R,3R,4R,5R,6R)-5-hydroxy-3,4-dimethoxy-6-methyltetrahydro-2H-pyran-2-yl)oxy)propan-2-yl)-10-(((2S,3R,4R,6R)-3-hydroxy-4-(methoxyamino)-6-methyltetrahydro-2H-pyran-2-yl)oxy)-3,5,7,9,11,13-hexamethyl-7-(((2-((2-nitrophenyl)sulfonamido)ethyl)carbamoyl)oxy)-6,14-dioxooxacyclotetradecan-4-yl 3-methylbutanoate, 16S rRNA, 23S rRNA, ... | | 著者 | Jenner, L.B, Yusupov, M, Yusupova, G. | | 登録日 | 2020-11-17 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Discovery of natural-product-derived sequanamycins as potent oral anti-tuberculosis agents.
Cell, 186, 2023
|
|
1GYZ
 
 | |
2FKX
 
 | |
6ELP
 
 | |
6EL5
 
 | | Estimation of relative drug-target residence times by random acceleration molecular dynamics simulation | | 分子名称: | 8-(2-CHLORO-3,4,5-TRIMETHOXY-BENZYL)-2-FLUORO-9-PENT-4-YLNYL-9H-PURIN-6-YLAMINE, Heat shock protein HSP 90-alpha | | 著者 | Musil, D, Lehmann, M, Eggenweiler, H.-M. | | 登録日 | 2017-09-27 | | 公開日 | 2018-05-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
6EI5
 
 | |
6ELO
 
 | |
