5Z8Z
 
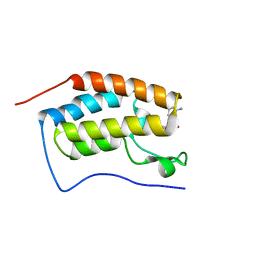 | | BRD4 Bromodomain 1 with an inhibitor | | Descriptor: | 6-[(3S)-3-azanylpiperidin-1-yl]sulfonyl-1-ethyl-benzo[cd]indol-2-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Chen, S, Chen, H. | | Deposit date: | 2018-02-01 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | BRD4 Bromodomain 1 with an inhibitor
To Be Published
|
|
5ZND
 
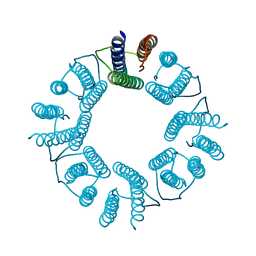 | | 8-mer nanotube derived from 24-mer rHuHF nanocage | | Descriptor: | Ferritin heavy chain | | Authors: | Wang, W.M, Wang, L.L, Zang, J.C, Chen, H, Zhao, G.H, Wang, H.F. | | Deposit date: | 2018-04-09 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Selective Elimination of the Key Subunit Interfaces Facilitates Conversion of Native 24-mer Protein Nanocage into 8-mer Nanorings.
J. Am. Chem. Soc., 140, 2018
|
|
7EVP
 
 | | Cryo-EM structure of the Gp168-beta-clamp complex | | Descriptor: | Beta sliding clamp, Sliding clamp inhibitor | | Authors: | Liu, B, Li, S, Liu, Y, Chen, H, Hu, Z, Wang, Z, Gou, L, Zhang, L, Ma, B, Wang, H, Matthews, S, Wang, Y, Zhang, K. | | Deposit date: | 2021-05-21 | | Release date: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Bacteriophage Twort protein Gp168 is a beta-clamp inhibitor by occupying the DNA sliding channel.
Nucleic Acids Res., 49, 2021
|
|
6JYW
 
 | |
6M54
 
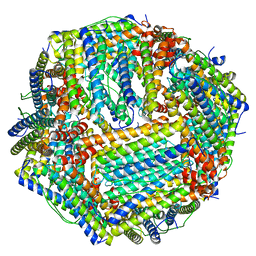 | | Human apo ferritin frozen on TEM grid with Amorphous nickel titanium alloy supporting film | | Descriptor: | FE (II) ION, Ferritin heavy chain | | Authors: | Huang, X, Zhang, L, Wen, Z, Chen, H, Li, S, Ji, G, Yin, C, Sun, F. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Amorphous nickel titanium alloy film: A new choice for cryo electron microscopy sample preparation.
Prog.Biophys.Mol.Biol., 156, 2020
|
|
6M52
 
 | | Human apo ferritin frozen on TEM grid with amorphous carbon supporting film | | Descriptor: | FE (II) ION, Ferritin heavy chain | | Authors: | Huang, X, Zhang, L, Wen, Z, Chen, H, Li, S, Ji, G, Yin, C, Sun, F. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Amorphous nickel titanium alloy film: A new choice for cryo electron microscopy sample preparation.
Prog.Biophys.Mol.Biol., 156, 2020
|
|
7V6G
 
 | | Structure of Candida albicans Fructose-1,6-bisphosphate aldolase mutation C157S with CN39 | | Descriptor: | 1,2-ETHANEDIOL, Fructose-bisphosphate aldolase, ZINC ION, ... | | Authors: | Cao, H, Huang, Y, Chen, H, Wan, C, Ren, Y, Wan, J. | | Deposit date: | 2021-08-20 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
8IMQ
 
 | |
7WPY
 
 | | AndA_M119A_N121V variant | | Descriptor: | Dioxygenase andA, FE (III) ION | | Authors: | Mori, T, Chen, H, Abe, I. | | Deposit date: | 2022-01-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | AndA_M119A_N121V variant
To Be Published
|
|
6K9H
 
 | | Human LXR-beta in complex with an agonist | | Descriptor: | Oxysterols receptor LXR-beta, ~{tert}-butyl (2'~{S},3~{S})-2-oxidanylidene-2'-phenyl-spiro[1~{H}-indole-3,3'-pyrrolidine]-1'-carboxylate | | Authors: | Zhang, Z, Zhou, H. | | Deposit date: | 2019-06-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of new LXR beta agonists as glioblastoma inhibitors.
Eur.J.Med.Chem., 194, 2020
|
|
8EX8
 
 | |
8EX6
 
 | | Human S1P transporter Spns2 in an inward-facing open conformation (state 1*) | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Ahmed, S, Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural and functional insights into Spns2-mediated transport of sphingosine-1-phosphate.
Cell, 186, 2023
|
|
8EX4
 
 | | Human S1P transporter Spns2 in an inward-facing open conformation (state 1) | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Ahmed, S, Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural and functional insights into Spns2-mediated transport of sphingosine-1-phosphate.
Cell, 186, 2023
|
|
8EX5
 
 | |
8EX7
 
 | |
6XES
 
 | | Tubulin-RB3_SLD in complex with compound 40a | | Descriptor: | DIMETHYL SULFOXIDE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-06-13 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Stable Colchicine-Binding Site Tubulin Inhibitors 6-Aryl-2-benzoyl-pyridines as Potential Anticancer Agents.
J.Med.Chem., 64, 2021
|
|
6XER
 
 | | Tubulin-RB3_SLD in complex with colchicine | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-06-13 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Stable Colchicine-Binding Site Tubulin Inhibitors 6-Aryl-2-benzoyl-pyridines as Potential Anticancer Agents.
J.Med.Chem., 64, 2021
|
|
6XET
 
 | | Tubulin-RB3_SLD in complex with compound 60c | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-06-13 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Stable Colchicine-Binding Site Tubulin Inhibitors 6-Aryl-2-benzoyl-pyridines as Potential Anticancer Agents.
J.Med.Chem., 64, 2021
|
|
6K9G
 
 | | Human LXR-beta in complex with an agonist | | Descriptor: | Oxysterols receptor LXR-beta, ~{tert}-butyl (2'~{R},3~{R})-2'-[3-[4-(hydroxymethyl)-3-methylsulfonyl-phenyl]phenyl]-2-oxidanylidene-spiro[1~{H}-indole-3,3'-pyrrolidine]-1'-carboxylate | | Authors: | Zhang, Z, Zhou, H. | | Deposit date: | 2019-06-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of new LXR beta agonists as glioblastoma inhibitors.
Eur.J.Med.Chem., 194, 2020
|
|
6PC4
 
 | | Tubulin-RB3_SLD-TTL in complex with compound ABI-274 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2019-06-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structure-Activity Relationship Study of Novel 6-Aryl-2-benzoyl-pyridines as Tubulin Polymerization Inhibitors with Potent Antiproliferative Properties.
J.Med.Chem., 63, 2020
|
|
7VRA
 
 | | The crystal structure of EGFR T790M/C797S with the inhibitor HC5476 | | Descriptor: | 25-chloro-11-(ethylsulfonyl)-44-morpholino-11H-5,12-dioxa-3-aza-1(3,6)-indola-2(4,2)-pyrimidina-4(1,3)-benzenacyclododecaphane, Epidermal growth factor receptor | | Authors: | Zhu, S.J. | | Deposit date: | 2021-10-22 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Conformational Constrained 4-(1-Sulfonyl-3-indol)yl-2-phenylaminopyrimidine Derivatives as New Fourth-Generation Epidermal Growth Factor Receptor Inhibitors Targeting T790M/C797S Mutations.
J.Med.Chem., 65, 2022
|
|
7VRE
 
 | | The crystal structure of EGFR T790M/C797S with the inhibitor HCD2892 | | Descriptor: | 5-chloranyl-N-[5-chloranyl-2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]-4-(1-ethylsulfonylindol-3-yl)pyrimidin-2-amine, Epidermal growth factor receptor | | Authors: | Zhu, S.J. | | Deposit date: | 2021-10-22 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Conformational Constrained 4-(1-Sulfonyl-3-indol)yl-2-phenylaminopyrimidine Derivatives as New Fourth-Generation Epidermal Growth Factor Receptor Inhibitors Targeting T790M/C797S Mutations.
J.Med.Chem., 65, 2022
|
|
4ZSG
 
 | | MITOGEN ACTIVATED PROTEIN KINASE 7 IN COMPLEX WITH INHIBITOR | | Descriptor: | 3-amino-5-[(4-chlorophenyl)amino]-N-(propan-2-yl)-1H-1,2,4-triazole-1-carboxamide, GLYCEROL, Mitogen-activated protein kinase 7 | | Authors: | Tucker, J, Ogg, D.J. | | Deposit date: | 2015-05-13 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4ZSJ
 
 | | MITOGEN ACTIVATED PROTEIN KINASE 7 IN COMPLEX WITH INHIBITOR | | Descriptor: | 3-amino-5-[(4-chloro-3-methylphenyl)amino]-N-(propan-2-yl)-1H-1,2,4-triazole-1-carboxamide, GLYCEROL, Mitogen-activated protein kinase 7 | | Authors: | Tucker, J, Ogg, D.J. | | Deposit date: | 2015-05-13 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4ZSL
 
 | | MITOGEN ACTIVATED PROTEIN KINASE 7 IN COMPLEX WITH INHIBITOR | | Descriptor: | 3-amino-5-[(4-chlorophenyl)amino]-N-[(1S)-1-phenylethyl]-1H-1,2,4-triazole-1-carboxamide, GLYCEROL, Mitogen-activated protein kinase 7 | | Authors: | Ogg, D.J, Tucker, J. | | Deposit date: | 2015-05-13 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
