2MPV
 
 | | Structural insight into host recognition and biofilm formation by aggregative adherence fimbriae of enteroaggregative Esherichia coli | | Descriptor: | Major fimbrial subunit of aggregative adherence fimbria II AafA | | Authors: | Matthews, S.J, Yang, Y, Berry, A.A, Pakharukova, N, Garnett, J.A, Lee, W, Cota, E, Liu, B, Roy, S, Tuittila, M, Marchant, J, Inman, K.G, Ruiz-Perez, F, Mandomando, I, Nataro, J.P, Zavialov, A.V. | | Deposit date: | 2014-06-04 | | Release date: | 2014-10-29 | | Method: | SOLUTION NMR | | Cite: | Structural insight into host recognition by aggregative adherence fimbriae of enteroaggregative Escherichia coli.
Plos Pathog., 10, 2014
|
|
1TAM
 
 | | HUMAN IMMUNODEFICIENCY VIRUS, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HIV-1 MATRIX PROTEIN | | Authors: | Matthews, S, Barlow, P, Clark, N, Kingsman, S, Kingsman, A, Campbell, I. | | Deposit date: | 1996-02-07 | | Release date: | 1996-07-11 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of p17, the HIV matrix protein.
Biochem.Soc.Trans., 23, 1995
|
|
2K2S
 
 | |
6Y1H
 
 | |
5O65
 
 | | Crystal Structure of the Pseudomonas functional amyloid secretion protein FapF | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, FapF, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Rouse, S.L, Hare, S, Lambert, S, Morgan, R.M.L, Hawthorne, W.J, Berry, J, Matthews, S.J. | | Deposit date: | 2017-06-05 | | Release date: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new class of hybrid secretion system is employed in Pseudomonas amyloid biogenesis.
Nat Commun, 8, 2017
|
|
5O67
 
 | | Crystal structure of the FapF polypeptide transporter - F103A mutant | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, FapF, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Rouse, S.L, Hare, S, Lambert, S, Morgan, R.M.L, Hawthorne, W.J, Berry, J, Matthews, S.J. | | Deposit date: | 2017-06-05 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | A new class of hybrid secretion system is employed in Pseudomonas amyloid biogenesis.
Nat Commun, 8, 2017
|
|
5O68
 
 | | Crystal Structure of the Pseudomonas functional amyloid secretion protein FapF - R157A mutant | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, FapF, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Rouse, S.L, Hare, S, Lambert, S, Morgan, R.M.L, Hawthorne, W.J, Berry, J, Matthews, S.J. | | Deposit date: | 2017-06-05 | | Release date: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | A new class of hybrid secretion system is employed in Pseudomonas amyloid biogenesis.
Nat Commun, 8, 2017
|
|
5LG9
 
 | |
5AAY
 
 | | TBK1 recruitment to cytosol-invading Salmonella induces anti- bacterial autophagy | | Descriptor: | NF-KAPPA-B ESSENTIAL MODULATOR, ZINC ION | | Authors: | Thurston, T.l, Allen, M.D, Ravenhill, B, Karpiyevitch, M, Bloor, S, Kaul, A, Matthews, S, Komander, D, Holden, D, Bycroft, M, Randow, F. | | Deposit date: | 2015-07-31 | | Release date: | 2016-07-13 | | Last modified: | 2019-10-23 | | Method: | SOLUTION NMR | | Cite: | Recruitment of Tbk1 to Cytosol-Invading Salmonella Induces Wipi2-Dependent Antibacterial Autophagy.
Embo J., 35, 2016
|
|
5AAZ
 
 | | TBK1 recruitment to cytosol-invading Salmonella induces anti- bacterial autophagy | | Descriptor: | OPTINEURIN, ZINC ION | | Authors: | Thurston, T.l, Allen, M.D, Ravenhill, B, Karpiyevitch, M, Bloor, S, Kaul, A, Matthews, S, Komander, D, Holden, D, Bycroft, M, Randow, F. | | Deposit date: | 2015-07-31 | | Release date: | 2016-07-13 | | Last modified: | 2016-08-31 | | Method: | SOLUTION NMR | | Cite: | Recruitment of Tbk1 to Cytosol-Invading Salmonella Induces Wipi2-Dependent Antibacterial Autophagy.
Embo J., 35, 2016
|
|
5AAQ
 
 | | TBK1 recruitment to cytosol-invading Salmonella induces anti- bacterial autophagy | | Descriptor: | CALCIUM-BINDING AND COILED-COIL DOMAIN-CONTAINING PROTEIN 2, ZINC ION | | Authors: | Thurston, T.L, Allen, M.D, Ravenhill, B, Karpiyevitch, M, Bloor, S, Kaul, A, Matthews, S, Komander, D, Holden, D, Bycroft, M, Randow, F. | | Deposit date: | 2015-07-28 | | Release date: | 2016-07-13 | | Last modified: | 2019-10-23 | | Method: | SOLUTION NMR | | Cite: | Recruitment of Tbk1 to Cytosol-Invading Salmonella Induces Wipi2-Dependent Antibacterial Autophagy.
Embo J., 35, 2016
|
|
5LGM
 
 | | Gp5.7 mutant L42A | | Descriptor: | Fusion protein 5.5/5.7 | | Authors: | Liu, B, Matthews, S. | | Deposit date: | 2016-07-07 | | Release date: | 2017-08-16 | | Method: | SOLUTION NMR | | Cite: | Gp5.7 mutant L42A
to be published
|
|
4XZS
 
 | | Crystal Structure of TRIAP1-MBP fusion | | Descriptor: | Maltose-binding periplasmic protein,TP53-regulated inhibitor of apoptosis 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Miliara, X, Garnett, J.A, Abid-Ali, F, Perez-Dorado, I, Matthews, S.J. | | Deposit date: | 2015-02-04 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural insight into the TRIAP1/PRELI-like domain family of mitochondrial phospholipid transfer complexes.
Embo Rep., 16, 2015
|
|
4XZV
 
 | | Crystal Structure of SLMO1-TRIAP1 Complex | | Descriptor: | Maltose-binding periplasmic protein,TP53-regulated inhibitor of apoptosis 1, Protein slowmo homolog 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Miliara, X, Garnett, J.A, Matthews, S.J. | | Deposit date: | 2015-02-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Structural insight into the TRIAP1/PRELI-like domain family of mitochondrial phospholipid transfer complexes.
Embo Rep., 16, 2015
|
|
8AG0
 
 | | Crystal structure of mutant PRELID3a-TRIAP1 complex - R53E | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,TP53-regulated inhibitor of apoptosis 1, PRELI domain containing protein 3A, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Milara, X, Perez-Dorado, J.I, Matthews, S.J. | | Deposit date: | 2022-07-18 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An intermolecular hydrogen bonded network in the PRELID-TRIAP protein family plays a role in lipid sensing.
Biochim Biophys Acta Proteins Proteom, 1871, 2022
|
|
5M0O
 
 | | Crystal structure of cytochrome P450 OleT H85Q in complex with arachidonic acid | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Tee, K.L, Munro, A, Matthews, S, Leys, D, Levy, C. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-11 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic Determinants of Alkene Production by the Cytochrome P450 Peroxygenase OleTJE.
J. Biol. Chem., 292, 2017
|
|
5M0N
 
 | | Crystal structure of cytochrome P450 OleT in complex with formate | | Descriptor: | FORMIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Tee, K.L, Munro, A, Matthews, S, Leys, D, Levy, C. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Catalytic Determinants of Alkene Production by the Cytochrome P450 Peroxygenase OleTJE.
J. Biol. Chem., 292, 2017
|
|
6TXT
 
 | |
5M0P
 
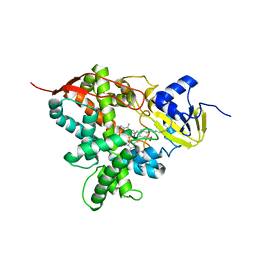 | | Crystal structure of cytochrome P450 OleT F79A in complex with arachidonic acid | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, Terminal olefin-forming fatty acid decarboxylase, ... | | Authors: | Tee, K.L, Munro, A, Matthews, S, Leys, D, Levy, C. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-11 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Catalytic Determinants of Alkene Production by the Cytochrome P450 Peroxygenase OleTJE.
J. Biol. Chem., 292, 2017
|
|
8C4A
 
 | |
7EVP
 
 | | Cryo-EM structure of the Gp168-beta-clamp complex | | Descriptor: | Beta sliding clamp, Sliding clamp inhibitor | | Authors: | Liu, B, Li, S, Liu, Y, Chen, H, Hu, Z, Wang, Z, Gou, L, Zhang, L, Ma, B, Wang, H, Matthews, S, Wang, Y, Zhang, K. | | Deposit date: | 2021-05-21 | | Release date: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Bacteriophage Twort protein Gp168 is a beta-clamp inhibitor by occupying the DNA sliding channel.
Nucleic Acids Res., 49, 2021
|
|
3QS3
 
 | |
3QS2
 
 | |
5LVY
 
 | |
3RGU
 
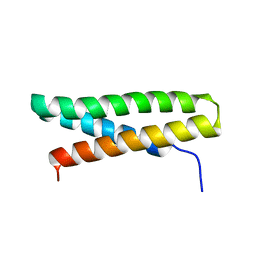 | | Structure of Fap-NRa at pH 5.0 | | Descriptor: | Fimbriae-associated protein Fap1, alpha-D-glucopyranose | | Authors: | Garnett, J.A, Matthews, S.J. | | Deposit date: | 2011-04-09 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insight into the role of Streptococcus parasanguinis Fap1 within oral biofilm formation.
Biochem.Biophys.Res.Commun., 417, 2012
|
|
