4HZ5
 
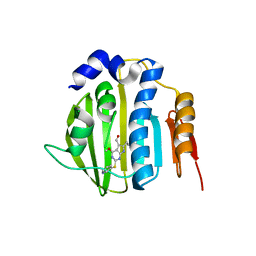 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity | | Descriptor: | 6-ethyl-4-methoxy-2-(pyridin-3-ylsulfanyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbaldehyde, DNA topoisomerase IV, B subunit | | Authors: | Bensen, D.C, Creighton, C.J, Tari, L.W. | | Deposit date: | 2012-11-14 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4DUS
 
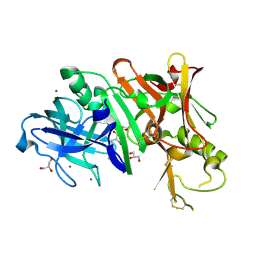 | | Structure of Bace-1 (Beta-Secretase) in complex with N-((2S,3R)-1-(4-fluorophenyl)-3-hydroxy-4-((6'-neopentyl-3',4'-dihydrospiro[cyclobutane-1,2'-pyrano[2,3-b]pyridin]-4'-yl)amino)butan-2-yl)acetamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Sickmier, E.A. | | Deposit date: | 2012-02-22 | | Release date: | 2012-10-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Potent and Orally Efficacious, Hydroxyethylamine-Based Inhibitor of beta-Secretase.
ACS Med Chem Lett, 3, 2012
|
|
4DCH
 
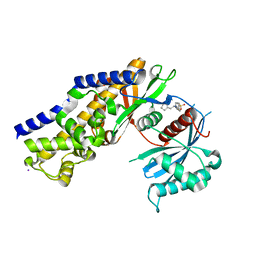 | | Insights into Glucokinase Activation Mechanism: Observation of Multiple Distinct Protein Conformations | | Descriptor: | (2R)-3-cyclopentyl-2-[4-(methylsulfonyl)phenyl]-N-(1,3-thiazol-2-yl)propanamide, Glucokinase, IODIDE ION, ... | | Authors: | Greasley, S.E, Hickey, M, Feng, J, Garcia, E. | | Deposit date: | 2012-01-17 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Insights into Mechanism of Glucokinase Activation: OBSERVATION OF MULTIPLE DISTINCT PROTEIN CONFORMATIONS.
J.Biol.Chem., 287, 2012
|
|
8GRA
 
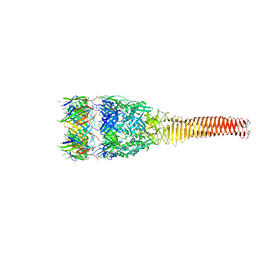 | |
7E7E
 
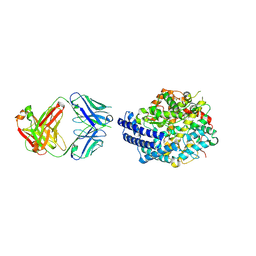 | | The co-crystal structure of ACE2 with Fab | | Descriptor: | Processed angiotensin-converting enzyme 2, ZINC ION, h11B11-Fab | | Authors: | Xiao, J.Y, Zhang, Y. | | Deposit date: | 2021-02-26 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A broadly neutralizing humanized ACE2-targeting antibody against SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7UBA
 
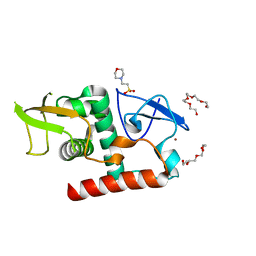 | | Structure of fungal Hop1 CBR domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HORMA domain-containing protein, PENTAETHYLENE GLYCOL, ... | | Authors: | Ur, S.N, Corbett, K.D. | | Deposit date: | 2022-03-14 | | Release date: | 2023-03-29 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Chromatin binding by HORMAD proteins regulates meiotic recombination initiation.
Embo J., 43, 2024
|
|
6S50
 
 | |
5C91
 
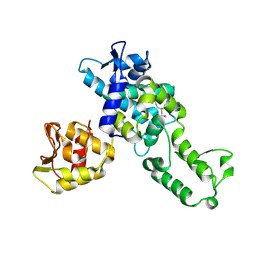 | | NEDD4 HECT with covalently bound indole-based inhibitor | | Descriptor: | E3 ubiquitin-protein ligase NEDD4, methyl (2E)-4-{[(5-methoxy-1,2-dimethyl-1H-indol-3-yl)carbonyl]amino}but-2-enoate | | Authors: | Span, I, Smith, A.T, Kathman, S, Statsyuk, A.V, Rosenzweig, A.C. | | Deposit date: | 2015-06-26 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | A Small Molecule That Switches a Ubiquitin Ligase From a Processive to a Distributive Enzymatic Mechanism.
J. Am. Chem. Soc., 137, 2015
|
|
3RL7
 
 | | Crystal structure of hDLG1-PDZ1 complexed with APC | | Descriptor: | 11-mer peptide from Adenomatous polyposis coli protein, Disks large homolog 1 | | Authors: | Zhang, Z, Li, H, Wu, G. | | Deposit date: | 2011-04-19 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the recognition of adenomatous polyposis coli by the Discs Large 1 protein.
Plos One, 6, 2011
|
|
3RL8
 
 | | Crystal structure of hDLG1-PDZ2 complexed with APC | | Descriptor: | 11-mer peptide from Adenomatous polyposis coli protein, Disks large homolog 1 | | Authors: | Zhang, Z, Li, H, Wu, G. | | Deposit date: | 2011-04-19 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for the recognition of adenomatous polyposis coli by the Discs Large 1 protein.
Plos One, 6, 2011
|
|
4MC7
 
 | | Crystal structure of a subtype N11 neuraminidase-like protein of A/flat-faced bat/Peru/033/2010 (H18N11) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Stevens, J. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | New world bats harbor diverse influenza a viruses.
Plos Pathog., 9, 2013
|
|
8GBL
 
 | | Structure of Apo Human SIRT5 | | Descriptor: | NAD-dependent protein deacylase sirtuin-5, mitochondrial, ZINC ION | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2023-02-26 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Human SIRT5 variants with reduced stability and activity do not cause neuropathology in mice.
Iscience, 27, 2024
|
|
8GBN
 
 | | Structure of Apo Human SIRT5 P114T Mutant | | Descriptor: | 1,2-ETHANEDIOL, NAD-dependent protein deacylase sirtuin-5, mitochondrial, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2023-02-26 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human SIRT5 variants with reduced stability and activity do not cause neuropathology in mice.
Iscience, 27, 2024
|
|
8H69
 
 | | Cryo-EM structure of influenza RNA polymerase | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*UP*AP*AP*AP*CP*UP*CP*CP*UP*GP*CP*UP*UP*UP*UP*GP*CP*U)-3'), ... | | Authors: | Li, H, Wu, Y, Liang, H, Liu, Y. | | Deposit date: | 2022-10-16 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An intermediate state allows influenza polymerase to switch smoothly between transcription and replication cycles.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8GVK
 
 | |
7D3I
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with MI-23 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[3-[3,5-bis(fluoranyl)phenyl]propanoyl]-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase | | Authors: | Zeng, R, Li, Y.S, Qiao, J.X, Wang, Y.F, Yang, S.Y, Lei, J. | | Deposit date: | 2020-09-19 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
4K3Y
 
 | |
6BMK
 
 | | Crystal structure of MHC-I like protein | | Descriptor: | (2R)-1-(decanoyloxy)-3-(phosphonooxy)propan-2-yl octadecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Khandokar, Y.B, Le Nours, J, Rossjohn, J. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Differing roles of CD1d2 and CD1d1 proteins in type I natural killer T cell development and function.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4JSC
 
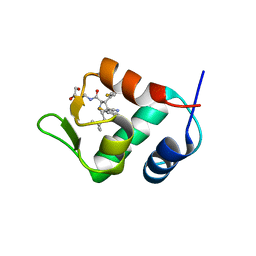 | | The 2.5A crystal structure of humanized Xenopus MDM2 with RO5316533 - a pyrrolidine MDM2 inhibitor | | Descriptor: | (3S,4R,5S)-3-(3-chloro-2-fluorophenyl)-4-(4-chloro-2-fluorophenyl)-4-cyano-N-[(3S)-3,4-dihydroxybutyl]-5-(2,2-dimethylpropyl)-D-prolinamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Janson, C.A, Lukacs, C, Graves, B. | | Deposit date: | 2013-03-22 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of RG7388, a Potent and Selective p53-MDM2 Inhibitor in Clinical Development.
J.Med.Chem., 56, 2013
|
|
8GVL
 
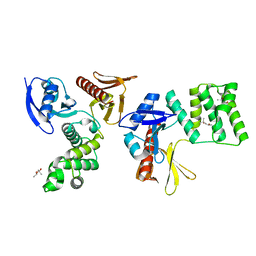 | | PTPN21 FERM | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Tyrosine-protein phosphatase non-receptor type 21 | | Authors: | Chen, L, Zheng, Y.Y, Zhou, C. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of PTPN21 reveals a dominant-negative effect of the FERM domain on its phosphatase activity.
Sci Adv, 10, 2024
|
|
8GWH
 
 | |
8GVV
 
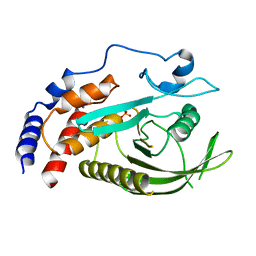 | | PTPN21 PTP domain C1108S mutant | | Descriptor: | IODIDE ION, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 21 | | Authors: | Chen, L, Zheng, Y.Y, Zhou, C. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of PTPN21 reveals a dominant-negative effect of the FERM domain on its phosphatase activity.
Sci Adv, 10, 2024
|
|
8HBT
 
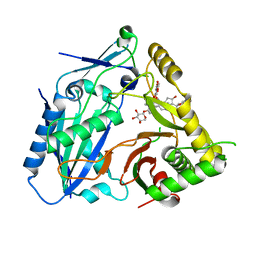 | | AmAT7-3 mutant A310G | | Descriptor: | AmAT7-3-A310G, Astragaloside IV | | Authors: | Wang, L.L. | | Deposit date: | 2022-10-31 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Characterization and structure-based protein engineering of a regiospecific saponin acetyltransferase from Astragalus membranaceus.
Nat Commun, 14, 2023
|
|
8GXE
 
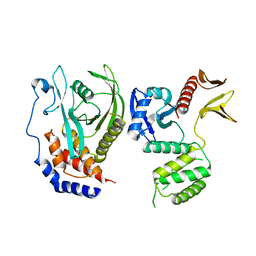 | | PTPN21 FERM PTP complex | | Descriptor: | CHLORIDE ION, Tyrosine-protein phosphatase non-receptor type 21 | | Authors: | Chen, L, Zheng, Y.Y, Zhou, C. | | Deposit date: | 2022-09-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of PTPN21 reveals a dominant-negative effect of the FERM domain on its phosphatase activity.
Sci Adv, 10, 2024
|
|
8H8I
 
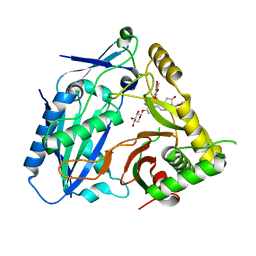 | | Triterpenoid saponin acetyltransferase, AmAT7-3 | | Descriptor: | AmAT7-3, Astragaloside IV | | Authors: | Wang, L.L. | | Deposit date: | 2022-10-22 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Characterization and structure-based protein engineering of a regiospecific saponin acetyltransferase from Astragalus membranaceus.
Nat Commun, 14, 2023
|
|
