6UFX
 
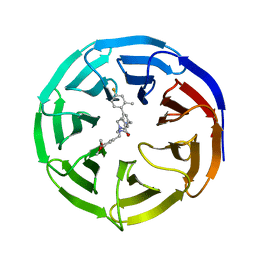 | | WD repeat-containing protein 5 complexed with N-[(3,5-dimethoxyphenyl)methyl]-4'-fluoro-5-{[(2E)-2-imino-3-methyl-2,3-dihydro-1H-imidazol-1-yl]methyl}-2'-methyl[1,1'-biphenyl]-3-carboxamide (compound 13) | | Descriptor: | N-[(3,5-dimethoxyphenyl)methyl]-4'-fluoro-5-{[(2E)-2-imino-3-methyl-2,3-dihydro-1H-imidazol-1-yl]methyl}-2'-methyl[1,1'-biphenyl]-3-carboxamide, WD repeat-containing protein 5 | | Authors: | Rietz, T.A, Fesik, S.W, Zhao, B. | | Deposit date: | 2019-09-25 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Discovery and Structure-Based Optimization of Potent and Selective WD Repeat Domain 5 (WDR5) Inhibitors Containing a Dihydroisoquinolinone Bicyclic Core.
J.Med.Chem., 63, 2020
|
|
4Z8Q
 
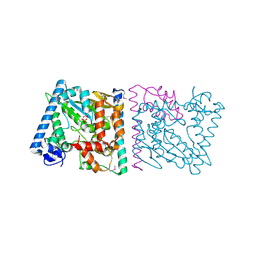 | | CRYSTAL STRUCTURE OF AvrRxo1-ORF1:AvrRxo1-ORF2 COMPLEX, SELENOMETHIONINE SUBSTITUTED. | | Descriptor: | AvrRxo1-ORF1, AvrRxo1-ORF2, PHOSPHATE ION | | Authors: | Han, Q, Zhou, C, Wu, S, Liu, Y, Yang, Z, Miao, J, Triplett, L, Cheng, Q, Tokuhisa, J, Deblais, L, Robinson, H, Leach, J.E, Li, J, Zhao, B. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Xanthomonas AvrRxo1-ORF1, a Type III Effector with a Polynucleotide Kinase Domain, and Its Interactor AvrRxo1-ORF2.
Structure, 23, 2015
|
|
4Z8V
 
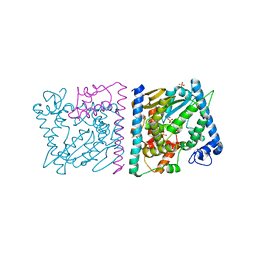 | | CRYSTAL STRUCTURE OF AVRRXO1-ORF1:-ORF2 COMPLEX, NATIVE. | | Descriptor: | AvrRxo1-ORF1, AvrRxo1-ORF2, PHOSPHATE ION | | Authors: | Han, Q, Zhou, C, Wu, S, Liu, Y, Yang, Z, Miao, J, Triplett, L, Cheng, Q, Tokuhisa, J, Deblais, L, Robinson, H, Leach, J.E, Li, J, Zhao, B. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Xanthomonas AvrRxo1-ORF1, a Type III Effector with a Polynucleotide Kinase Domain, and Its Interactor AvrRxo1-ORF2.
Structure, 23, 2015
|
|
4Z8T
 
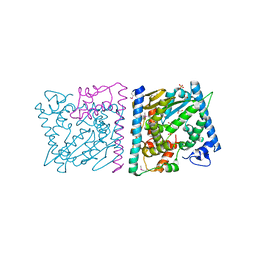 | | CRYSTAL STRUCTURE OF AvrRxo1-ORF1:AvrRxo1-ORF2 WITH SULPHATE IONS | | Descriptor: | ACETATE ION, AvrRxo1-ORF1, AvrRxo1-ORF2, ... | | Authors: | Han, Q, Zhou, C, Wu, S, Liu, Y, Yang, Z, Miao, J, Triplett, L, Cheng, Q, Tokuhisa, J, Deblais, L, Robinson, H, Leach, J.E, Li, J, Zhao, B. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure of Xanthomonas AvrRxo1-ORF1, a Type III Effector with a Polynucleotide Kinase Domain, and Its Interactor AvrRxo1-ORF2.
Structure, 23, 2015
|
|
6V43
 
 | | Crystal structure of the flavin oxygenase with cofactor and substrate bound involved in folate catabolism | | Descriptor: | FAD/FMN-containing dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, pteridine-2,4(1H,3H)-dione | | Authors: | Begley, T.P, Adak, S, Zhao, B, Li, P. | | Deposit date: | 2019-11-27 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A novel flavoenzyme catalyzed Baeyer-Villiger type rearrangement in bacterial folic acid catabolic pathway
To Be Published
|
|
6V42
 
 | |
4X0W
 
 | | The crystal structure of mupain-1-17 in complex with murinised human uPA | | Descriptor: | SULFATE ION, Urokinase-type plasminogen activator, mupain-1-17, ... | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-24 | | Release date: | 2015-10-21 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinctive binding modes and inhibitory mechanisms of two peptidic inhibitors of urokinase-type plasminogen activator with isomeric P1 residues.
Int.J.Biochem.Cell Biol., 62, 2015
|
|
4X1P
 
 | | The crystal structure of mupain-1-17 in complex with murinised human uPA at pH4.6 | | Descriptor: | MUPAIN-1-17, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-10-21 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Distinctive binding modes and inhibitory mechanisms of two peptidic inhibitors of urokinase-type plasminogen activator with isomeric P1 residues.
Int.J.Biochem.Cell Biol., 62, 2015
|
|
5UL6
 
 | | The molecular mechanisms by which NS1 of the 1918 Spanish influenza A virus hijack host protein-protein interactions | | Descriptor: | Adapter molecule crk, Proline-rich motif of nonstructural protein 1 of influenza a virus | | Authors: | Shen, Q, Zeng, D, Zhao, B, Li, P, Cho, J.H. | | Deposit date: | 2017-01-24 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Molecular Mechanisms Underlying the Hijack of Host Proteins by the 1918 Spanish Influenza Virus.
ACS Chem. Biol., 12, 2017
|
|
5W4Y
 
 | | Crystal Structure of Riboflavin Lyase (RcaE) with cofactor FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Riboflavin Lyase | | Authors: | Bhandari, D.M, Chakrabarty, Y, Zhao, B, Wood, J, Li, P, Begley, T.P. | | Deposit date: | 2017-06-13 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cannibalism Among the Flavins: a Novel C-N Bond Cleavage in Riboflavin Catabolism Mediated by Flavin-Generated Superoxide Radical
To be Published
|
|
1SBG
 
 | |
5W4Z
 
 | | Crystal Structure of Riboflavin Lyase (RcaE) with modified FMN and substrate Riboflavin | | Descriptor: | 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)-3-O-phosphono-D-ribitol, RIBOFLAVIN, Riboflavin Lyase | | Authors: | Bhandari, D.M, Chakrabarty, Y, Zhao, B, Wood, J, Li, P, Begley, T.P. | | Deposit date: | 2017-06-13 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cannibalism Among the Flavins: a Novel C-N Bond Cleavage in Riboflavin Catabolism Mediated by Flavin-Generated Superoxide Radical
To be Published
|
|
6OX7
 
 | | The complex of 1918 NS1-ED and the iSH2 domain of the human p85beta subunit of PI3K | | Descriptor: | Non-structural protein 1, Phosphatidylinositol 3-kinase regulatory subunit beta | | Authors: | Shen, Q, Zhao, B, Li, P, Cho, J.H. | | Deposit date: | 2019-05-13 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular recognition of a host protein by NS1 of pandemic and seasonal influenza A viruses.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5W48
 
 | | Crystal Structure of Riboflavin Lyase (RcaE) | | Descriptor: | Riboflavin Lyase, SULFATE ION | | Authors: | Bhandari, D.M, Chakrabarty, Y, Zhao, B, Wood, J, Li, P, Begley, T.P. | | Deposit date: | 2017-06-09 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cannibalism Among the Flavins: a Novel C-N Bond Cleavage in Riboflavin Catabolism Mediated by Flavin-Generated Superoxide Radical
To be Published
|
|
5HG1
 
 | | Crystal Structure of Human Hexokinase 2 with cmpd 1, a C-2-substituted glucosamine | | Descriptor: | 2-deoxy-2-{[(2E)-3-(3,4-dichlorophenyl)prop-2-enoyl]amino}-alpha-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose, CITRATE ANION, ... | | Authors: | Campobasso, N, Zhao, B, Smallwood, A. | | Deposit date: | 2016-01-07 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5HFU
 
 | | Crystal Structure of Human Hexokinase 2 with cmpd 27, a 2-amido-6-benzenesulfonamide glucosamine | | Descriptor: | Hexokinase-2, ~{N}-[(2~{S},3~{R},4~{R},5~{S},6~{R})-6-[[(4-cyanophenyl)sulfonylamino]methyl]-2,4,5-tris(oxidanyl)oxan-3-yl]-3-phenyl-benzamide | | Authors: | Campobasso, N, Zhao, B, Smallwood, A. | | Deposit date: | 2016-01-07 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.923 Å) | | Cite: | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5HEX
 
 | | Crystal Structure of Human Hexokinase 2 with cmpd 30, a 2-amino-6-benzenesulfonamide glucosamine | | Descriptor: | 2-[(3-bromobenzene-1-carbonyl)amino]-6-{[(4-carboxy-5-methylfuran-2-yl)sulfonyl]amino}-2,6-dideoxy-alpha-D-glucopyranos e, Hexokinase-2 | | Authors: | Campobasso, N, Zhao, B, Smallwood, A. | | Deposit date: | 2016-01-06 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
1ZYS
 
 | | Co-crystal structure of Checkpoint Kinase Chk1 with a pyrrolo-pyridine inhibitor | | Descriptor: | N-{5-[4-(4-METHYLPIPERAZIN-1-YL)PHENYL]-1H-PYRROLO[2,3-B]PYRIDIN-3-YL}NICOTINAMIDE, SULFATE ION, Serine/threonine-protein kinase Chk1, ... | | Authors: | Stavenger, R.A, Zhao, B, Zhou, B.-B.S, Brown, M.J, Lee, D, Holt, D.A. | | Deposit date: | 2005-06-10 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pyrrolo[2,3-b]pyridines Inhibit the Checkpoint Kinase Chk1
To be Published
|
|
3KYS
 
 | | Crystal structure of human YAP and TEAD complex | | Descriptor: | 65 kDa Yes-associated protein, Transcriptional enhancer factor TEF-1 | | Authors: | Li, Z, Zhao, B, Wang, P, Chen, F, Dong, Z, Yang, H, Guan, K.L, Xu, Y. | | Deposit date: | 2009-12-07 | | Release date: | 2010-02-23 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the YAP and TEAD complex
Genes Dev., 24, 2010
|
|
3MAZ
 
 | | Crystal Structure of the Human BRDG1/STAP-1 SH2 Domain in Complex with the NTAL pTyr136 Peptide | | Descriptor: | CheD family protein, MALONATE ION, Signal-transducing adaptor protein 1 | | Authors: | Kaneko, T, Huang, H, Zhao, B, Li, L, Liu, H, Voss, C.K, Wu, C, Schiller, M.R, Li, S.S. | | Deposit date: | 2010-03-24 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Loops govern SH2 domain specificity by controlling access to binding pockets.
Sci.Signal., 3, 2010
|
|
1BY8
 
 | | THE CRYSTAL STRUCTURE OF HUMAN PROCATHEPSIN K | | Descriptor: | PROTEIN (PROCATHEPSIN K) | | Authors: | Lalonde, J.M, Zhao, B, Smith, W.W, Janson, C.A, Desjarlais, R.L, Tomaszek, T.A, Carr, T.J, Thompson, S.K, Yamashita, D.S, Veber, D.F, Abdel-Mequid, S.S. | | Deposit date: | 1998-10-27 | | Release date: | 1999-10-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of human procathepsin K.
Biochemistry, 38, 1999
|
|
1BP4
 
 | | USE OF PAPAIN AS A MODEL FOR THE STRUCTURE-BASED DESIGN OF CATHEPSIN K INHIBITORS. CRYSTAL STRUCTURES OF TWO PAPAIN INHIBITOR COMPLEXES DEMONSTRATE BINDING TO S'-SUBSITES. | | Descriptor: | N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide, PAPAIN | | Authors: | Lalonde, J.M, Zhao, B, Smith, W.W, Janson, C.A, Desjarlais, R.L, Tomaszek, T.A, Carr, T.J, Thompson, S.K, Yamashita, D.S, Veber, D.F, Abdel-Mequid, S.S. | | Deposit date: | 1998-08-12 | | Release date: | 1999-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Use of papain as a model for the structure-based design of cathepsin K inhibitors: crystal structures of two papain-inhibitor complexes demonstrate binding to S'-subsites.
J.Med.Chem., 41, 1998
|
|
1BQI
 
 | | USE OF PAPAIN AS A MODEL FOR THE STRUCTURE-BASED DESIGN OF CATHEPSIN K INHIBITORS. CRYSTAL STRUCTURES OF TWO PAPAIN INHIBITOR COMPLEXES DEMONSTRATE BINDING TO S'-SUBSITES. | | Descriptor: | CARBOBENZYLOXY-(L)-LEUCINYL-(L)LEUCINYL METHOXYMETHYLKETONE, PAPAIN | | Authors: | Lalonde, J.M, Zhao, B, Smith, W.W, Janson, C.A, Desjarlais, R.L, Tomaszek, T.A, Carr, T.J, Thompson, S.K, Yamashita, D.S, Veber, D.F, Abdel-Mequid, S.S. | | Deposit date: | 1998-08-16 | | Release date: | 1999-08-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Use of papain as a model for the structure-based design of cathepsin K inhibitors: crystal structures of two papain-inhibitor complexes demonstrate binding to S'-subsites.
J.Med.Chem., 41, 1998
|
|
4ROV
 
 | | The crystal structure of novel APOBEC3G CD2 head-to-tail dimer suggests the binding mode of full-length APOBEC3G to HIV-1 ssDNA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Lu, X, Zhang, T, Xu, Z, Liu, S, Zhao, B, Lan, W, Wang, C, Ding, J, Cao, C. | | Deposit date: | 2014-10-29 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of DNA cytidine deaminase ABOBEC3G catalytic deamination domain suggests a binding mode of full-length enzyme to single-stranded DNA
J.Biol.Chem., 290, 2015
|
|
4ROW
 
 | | The crystal structure of novel APOBEC3G CD2 head-to-tail dimer suggests the binding mode of full-length APOBEC3G to HIV-1 ssDNA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Lu, X, Zhang, T, Xu, Z, Liu, S, Zhao, B, Lan, W, Wang, C, Ding, J, Cao, C. | | Deposit date: | 2014-10-29 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of DNA cytidine deaminase ABOBEC3G catalytic deamination domain suggests a binding mode of full-length enzyme to single-stranded DNA
J.Biol.Chem., 290, 2015
|
|
