6D4I
 
 | | Crystal Structure of a Fc Fragment of Rhesus macaque (Macaca mulatta) IgG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Fc fragment of IgG2 | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | From Rhesus macaque to human: structural evolutionary pathways for immunoglobulin G subclasses.
Mabs, 11, 2019
|
|
5VK0
 
 | |
5VK1
 
 | |
8GD0
 
 | | Crystal Structure of HIV-1 LM/HT Clade A/E CRF01 GP120 Core in Complex with TFH-I-070-A6 | | Descriptor: | (3S)-N-(4-chloro-3-fluorophenyl)-1-(4-methylpiperazine-1-carbonyl)piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Nguyen, D.N, Pazgier, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Piperidine CD4-Mimetic Compounds Expose Vulnerable Env Epitopes Sensitizing HIV-1-Infected Cells to ADCC.
Viruses, 15, 2023
|
|
8GD1
 
 | | Crystal Structure of HIV-1 LM/HT Clade A/E CRF01 GP120 Core in Complex with ZXC-I-092 | | Descriptor: | (3S,5S)-5-(aminomethyl)-N-(4-chloro-3-fluorophenyl)-1-[(3R,5S)-3,4,5-trimethylpiperazine-1-carbonyl]piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Nguyen, D.N, Pazgier, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Piperidine CD4-Mimetic Compounds Expose Vulnerable Env Epitopes Sensitizing HIV-1-Infected Cells to ADCC.
Viruses, 15, 2023
|
|
8GJT
 
 | | Crystal Structure of HIV-1 LM/HT Clade A/E CRF01 GP120 Core in Complex with TFH-I-116-D1 | | Descriptor: | (3S)-N-(4-chloro-3-fluorophenyl)-1-[(3R,5S)-3,4,5-trimethylpiperazine-1-carbonyl]piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Nguyen, D.N, Pazgier, M. | | Deposit date: | 2023-03-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Piperidine CD4-Mimetic Compounds Expose Vulnerable Env Epitopes Sensitizing HIV-1-Infected Cells to ADCC.
Viruses, 15, 2023
|
|
8GD5
 
 | | Crystal Structure of HIV-1 LM/HT Clade A/E CRF01 GP120 Core in Complex with DL-I-102 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 LM/HS clade A/E CRF01 gp120 core, ... | | Authors: | Tolbert, W.D, Nguyen, D.N, Pazgier, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Piperidine CD4-Mimetic Compounds Expose Vulnerable Env Epitopes Sensitizing HIV-1-Infected Cells to ADCC.
Viruses, 15, 2023
|
|
8GD3
 
 | | Crystal Structure of HIV-1 LM/HT Clade A/E CRF01 GP120 Core in Complex with DL-I-101 | | Descriptor: | (3S,5R)-N-(4-chloro-3-fluorophenyl)-1-(4-glycylpiperazine-1-carbonyl)-5-(hydroxymethyl)piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Nguyen, D.N, Pazgier, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Piperidine CD4-Mimetic Compounds Expose Vulnerable Env Epitopes Sensitizing HIV-1-Infected Cells to ADCC.
Viruses, 15, 2023
|
|
4M0I
 
 | |
4P6X
 
 | | Crystal Structure of cortisol-bound glucocorticoid receptor ligand binding domain | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | Authors: | He, Y, Zhou, X.E, Tolbert, W.D, Powell, K, Melcher, K, Xu, H.E. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and mechanism for the design of highly potent glucocorticoids.
Cell Res., 24, 2014
|
|
4P6W
 
 | | Crystal Structure of mometasone furoate-bound glucocorticoid receptor ligand binding domain | | Descriptor: | Glucocorticoid receptor, MOMETASONE FUROATE, Nuclear receptor coactivator 2 | | Authors: | He, Y, Zhou, X.E, Tolbert, W.D, Powell, K, Melcher, K, Xu, H.E. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structures and mechanism for the design of highly potent glucocorticoids.
Cell Res., 24, 2014
|
|
1GG0
 
 | | CRYSTAL STRUCTURE ANALYSIS OF KDOP SYNTHASE AT 3.0 A | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE SYNTHASE, PHOSPHATE ION | | Authors: | Wagner, T, Kretsinger, R.H, Bauerle, R, Tolbert, W.D. | | Deposit date: | 2000-08-04 | | Release date: | 2000-10-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 3-Deoxy-D-manno-octulosonate-8-phosphate synthase from Escherichia coli. Model of binding of phosphoenolpyruvate and D-arabinose-5-phosphate.
J.Mol.Biol., 301, 2000
|
|
7RQ6
 
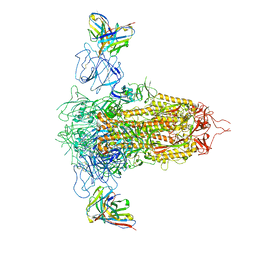 | | Cryo-EM structure of SARS-CoV-2 spike in complex with non-neutralizing NTD-directed CV3-13 Fab isolated from convalescent individual | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CV3-13 Fab heavy chain, ... | | Authors: | Chen, Y, Pozharski, E, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2021-08-05 | | Release date: | 2022-04-20 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | A Fc-enhanced NTD-binding non-neutralizing antibody delays virus spread and synergizes with a nAb to protect mice from lethal SARS-CoV-2 infection.
Cell Rep, 38, 2022
|
|
8GDJ
 
 | |
5DRZ
 
 | | Crystal structure of anti-HIV-1 antibody F240 Fab in complex with gp41 peptide | | Descriptor: | Envelope glycoprotein gp160, HIV Antibody F240 Heavy Chain, HIV Antibody F240 Light Chain, ... | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2015-09-16 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Molecular basis for epitope recognition by non-neutralizing anti-gp41 antibody F240.
Sci Rep, 6, 2016
|
|
8CTM
 
 | |
6MFJ
 
 | |
7NAB
 
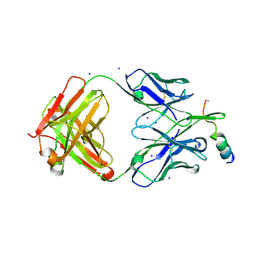 | | Crystal structure of human neutralizing mAb CV3-25 binding to SARS-CoV-2 S MPER peptide 1140-1165 | | Descriptor: | CITRIC ACID, CV3-25 Fab Heavy Chain, CV3-25 Fab Light Chain, ... | | Authors: | Chen, Y, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2021-06-21 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis and mode of action for two broadly neutralizing antibodies against SARS-CoV-2 emerging variants of concern.
Cell Rep, 38, 2022
|
|
7UL1
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with the neutralizing IGHV3-53-encoded antibody EH3 isolated from a nonvaccinated pediatric patient | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of EH3, Light chain of EH3, ... | | Authors: | Chen, Y, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2022-04-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD.
Iscience, 26, 2023
|
|
7UL0
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with the ridge-binding nAb EH8 isolated from a nonvaccinated pediatric patient | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of EH8, ... | | Authors: | Chen, Y, Tolbert, W.D, Bai, X, Pazgier, M. | | Deposit date: | 2022-04-03 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD.
Iscience, 26, 2023
|
|
7UOJ
 
 | |
4RFE
 
 | | Crystal structure of ADCC-potent ANTI-HIV-1 Rhesus macaque antibody JR4 Fab | | Descriptor: | CHLORIDE ION, Fab heavy chain of ADCC-potent anti-HIV-1 antibody JR4, Fab light chain of ADCC-potent anti-HIV-1 antibody JR4, ... | | Authors: | Wu, X, Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2014-09-25 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1.
J.Virol., 89, 2015
|
|
4RBW
 
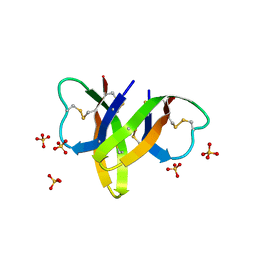 | | Crystal structure of human alpha-defensin 5, HD5 (Thr7Arg Glu21Arg mutant) | | Descriptor: | CHLORIDE ION, Defensin-5, SULFATE ION | | Authors: | Pazgier, M, Gohain, N, Tolbert, W.D. | | Deposit date: | 2014-09-13 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of a potent antibiotic peptide based on the active region of human defensin 5.
J.Med.Chem., 58, 2015
|
|
4RBX
 
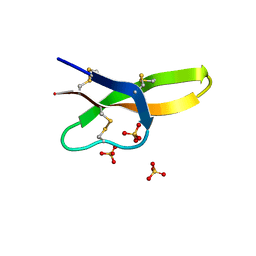 | |
4RFN
 
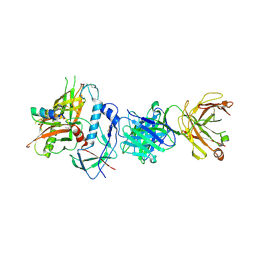 | | Crystal structure of ADCC-potent Rhesus macaque ANTIBODY JR4 in complex with HIV-1 CLADE A/E GP120 and M48 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB HEAVY CHAIN OF ADCC ANTI-HIV-1 ANTIBODY JR4, FAB LIGHT CHAIN OF ADCC ANTI-HIV-1 ANTIBODY JR4, ... | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2014-09-26 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1.
J.Virol., 89, 2015
|
|
