8T18
 
 | |
8T15
 
 | |
8T17
 
 | |
8SYG
 
 | |
7NSG
 
 | | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B | | Descriptor: | (+)-3-Hydroxy-4,5,6,6a-tetrahydro-3aH-pyrrolo[3,4-d]isoxazole-6-carboxylic acid, (-)-3-Hydroxy-4,5,6,6a-tetrahydro-3aH-pyrrolo[3,4-d]isoxazole-6-carboxylic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Baronina, A, Pike, A.C.W, Yu, X, Dong, Y.Y, Shintre, C.A, Tessitore, A, Chu, A, Rotty, B, Venkaya, S, Mukhopadhyay, S.M.M, Borkowska, O, Chalk, R, Shrestha, L, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Han, S, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-05 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B
TO BE PUBLISHED
|
|
6B5V
 
 | | Structure of TRPV5 in complex with econazole | | Descriptor: | 1-[(2R)-2-[(4-chlorobenzyl)oxy]-2-(2,4-dichlorophenyl)ethyl]-1H-imidazole, CALCIUM ION, Transient receptor potential cation channel subfamily V member 5 | | Authors: | Hughes, T.E.T, Lodowski, D.T, Huynh, K.W, Yazici, A, del Rosario, J, Kapoor, A, Basak, S, Samanta, A, Chakrapani, S, Zhou, Z.H, Filizola, M, Rohacs, T, Han, S, Moiseenkova-Bell, V.Y. | | Deposit date: | 2017-09-29 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of TRPV5 channel inhibition by econazole revealed by cryo-EM.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8E4Y
 
 | | Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with 2-oxohexadecyl-CoA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, Glycerol-3-phosphate acyltransferase 1, mitochondrial, ... | | Authors: | Johnson, Z.L, Wasilko, D.J, Ammirati, M, Chang, J.S, Han, S, Wu, H. | | Deposit date: | 2022-08-19 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of the acyl-transfer mechanism of human GPAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8E50
 
 | | Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with CoA and palmitoyl-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, COENZYME A, Glycerol-3-phosphate acyltransferase 1, ... | | Authors: | Wasilko, D.J, Johnson, Z.L, Ammirati, M, Chang, J.S, Han, S, Wu, H. | | Deposit date: | 2022-08-19 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural basis of the acyl-transfer mechanism of human GPAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7UJN
 
 | | Structure of Human SAMHD1 with Non-Hydrolysable dGTP Analog | | Descriptor: | 2'-deoxyguanosine-5'-O-(1-thiotriphosphate), Deoxynucleoside triphosphate triphosphohydrolase SAMHD1 | | Authors: | Huynh, K.W, Ammirati, M, Han, S. | | Deposit date: | 2022-03-31 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Phosphorylation of SAMHD1 Thr592 increases C-terminal domain dynamics, tetramer dissociation and ssDNA binding kinetics.
Nucleic Acids Res., 50, 2022
|
|
5ZBH
 
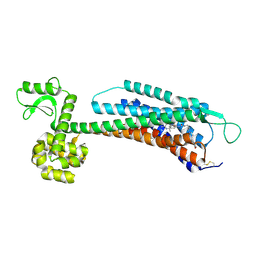 | | The Crystal Structure of Human Neuropeptide Y Y1 Receptor with BMS-193885 | | Descriptor: | Neuropeptide Y receptor type 1,T4 Lysozyme,Neuropeptide Y receptor type 1, dimethyl 4-{3-[({3-[4-(3-methoxyphenyl)piperidin-1-yl]propyl}carbamoyl)amino]phenyl}-2,6-dimethyl-1,4-dihydropyridine-3,5-dicarboxylate | | Authors: | Yang, Z, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2018-02-11 | | Release date: | 2018-04-25 | | Last modified: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of ligand binding modes at the neuropeptide Y Y1receptor
Nature, 556, 2018
|
|
5ZBQ
 
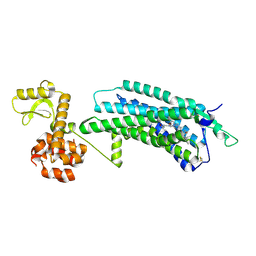 | | The Crystal Structure of human neuropeptide Y Y1 receptor with UR-MK299 | | Descriptor: | Neuropeptide Y receptor type 1,T4 Lysozyme, N~2~-(diphenylacetyl)-N-[(4-hydroxyphenyl)methyl]-N~5~-(N'-{[2-(propanoylamino)ethyl]carbamoyl}carbamimidoyl)-D-ornithinamide | | Authors: | Yang, Z, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2018-02-12 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand binding modes at the neuropeptide Y Y1receptor
Nature, 556, 2018
|
|
6KKU
 
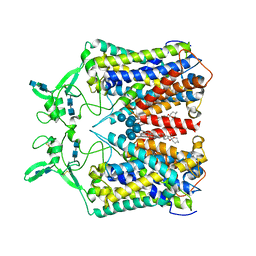 | | human KCC1 structure determined in NaCl and GDN | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Liu, S, Chang, S, Ye, S, Bai, X, Guo, J. | | Deposit date: | 2019-07-27 | | Release date: | 2019-10-23 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of the human cation-chloride cotransporter KCC1.
Science, 366, 2019
|
|
6KKT
 
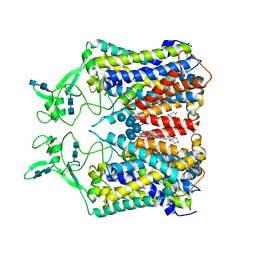 | | human KCC1 structure determined in KCl and lipid nanodisc | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Liu, S, Chang, S, Ye, S, Bai, X, Guo, J. | | Deposit date: | 2019-07-27 | | Release date: | 2019-10-23 | | Last modified: | 2020-08-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of the human cation-chloride cotransporter KCC1.
Science, 366, 2019
|
|
6KKR
 
 | | human KCC1 structure determined in KCl and detergent GDN | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Liu, S, Chang, S, Ye, S, Bai, X, Guo, J. | | Deposit date: | 2019-07-27 | | Release date: | 2019-10-23 | | Last modified: | 2020-08-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of the human cation-chloride cotransporter KCC1.
Science, 366, 2019
|
|
7S15
 
 | | GLP-1 receptor bound with Pfizer small molecule agonist | | Descriptor: | 2-[(4-{6-[(2,4-difluorophenyl)methoxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-[(1-ethyl-1H-imidazol-5-yl)methyl]-1H-benzimidazole-6-carboxylic acid, Glucagon-like peptide 1 receptor | | Authors: | Liu, Y, Dias, J.M, Han, S. | | Deposit date: | 2021-09-01 | | Release date: | 2022-06-08 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A Small-Molecule Oral Agonist of the Human Glucagon-like Peptide-1 Receptor.
J.Med.Chem., 65, 2022
|
|
5K18
 
 | | The NatB Acetyltransferase Complex Bound To bisubstrate inhibitor | | Descriptor: | Bisubstrate inhibitor, COENZYME A, N-terminal acetyltransferase B complex subunit NAT3, ... | | Authors: | Hong, H, Cai, Y, Zhang, S, Han, A. | | Deposit date: | 2016-05-17 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Molecular Basis of Substrate Specific Acetylation by N-Terminal Acetyltransferase NatB
Structure, 25, 2017
|
|
5K04
 
 | | The NatB Acetyltransferase Complex Bound To CoA and MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COENZYME A, N-terminal acetyltransferase B complex subunit NAT3, ... | | Authors: | Hong, H, Cai, Y, Zhang, S, Han, A. | | Deposit date: | 2016-05-17 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis of Substrate Specific Acetylation by N-Terminal Acetyltransferase NatB
Structure, 25, 2017
|
|
4ZQS
 
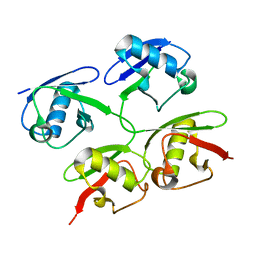 | | New compact conformation of linear Ub2 structure | | Descriptor: | ubiquitin | | Authors: | Thach, T.T, Shin, D, Han, S, Lee, S. | | Deposit date: | 2015-05-11 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | New conformations of linear polyubiquitin chains from crystallographic and solution-scattering studies expand the conformational space of polyubiquitin.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4XI3
 
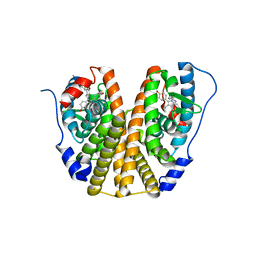 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with Bazedoxifene | | Descriptor: | Bazedoxifene, Estrogen receptor | | Authors: | Fanning, S.W, Mayne, C.G, Toy, W, Carlson, K, Greene, B, Nowak, J, Walter, R, Panchamukhi, S, Tajhorshid, E, Nettles, K.W, Chandarlapaty, S, Katzenellenbogen, J, Greene, G.L. | | Deposit date: | 2015-01-06 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | The SERM/SERD bazedoxifene disrupts ESR1 helix 12 to overcome acquired hormone resistance in breast cancer cells.
Elife, 7, 2018
|
|
4UCG
 
 | | NmeDAH7PS R126S variant | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, PHOSPHO-2-DEHYDRO-3-DEOXYHEPTONATE ALDOLASE, ... | | Authors: | Cross, P.J, Heyes, L.C, Zhang, S, Nazmi, A.R, Parker, E.J. | | Deposit date: | 2014-12-03 | | Release date: | 2016-01-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Functional Unit of Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase is Dimeric.
Plos One, 11, 2016
|
|
4V89
 
 | | Crystal Structure of Release Factor RF3 Trapped in the GTP State on a Rotated Conformation of the Ribosome (without viomycin) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhou, J, Lancaster, L, Trakhanov, S, Noller, H.F. | | Deposit date: | 2011-11-17 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome.
Rna, 18, 2012
|
|
4V63
 
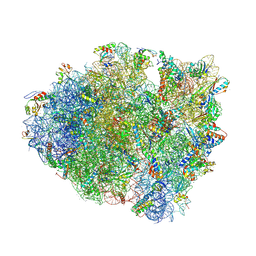 | | Structural basis for translation termination on the 70S ribosome. | | Descriptor: | 16S RRNA, 23S RRNA, 30S ribosomal protein S10, ... | | Authors: | Laurberg, M, Asahara, H, Korostelev, A, Zhu, J, Trakhanov, S, Noller, H.F. | | Deposit date: | 2008-05-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.207 Å) | | Cite: | Structural basis for translation termination on the 70S ribosome
Nature, 454, 2008
|
|
4V4J
 
 | | Interactions and Dynamics of the Shine-Dalgarno Helix in the 70S Ribosome. | | Descriptor: | 16S RNA, 23S LARGE SUBUNIT RIBOSOMAL RNA, 30S ribosomal protein S10, ... | | Authors: | Korostelev, A, Trakhanov, S, Asahara, H, Laurberg, M, Noller, H.F. | | Deposit date: | 2007-07-18 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.83 Å) | | Cite: | Interactions and dynamics of the Shine Dalgarno helix in the 70S ribosome.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3TUD
 
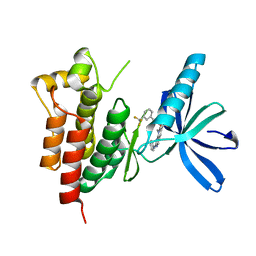 | | Crystal structure of SYK kinase domain with N-(4-methyl-3-(8-methyl-7-oxo-2-(phenylamino)-7,8-dihydropyrido[2,3-d]pyrimidin-6-yl)phenyl)-3-(trifluoromethyl)benzamide | | Descriptor: | N-{4-methyl-3-[8-methyl-7-oxo-2-(phenylamino)-7,8-dihydropyrido[2,3-d]pyrimidin-6-yl]phenyl}-3-(trifluoromethyl)benzamide, Tyrosine-protein kinase SYK | | Authors: | Lovering, F, McDonald, J, Whitlock, G, Glossop, P, Phillips, C, Sabnis, Y, Ryan, M, Fitz, L, Lee, J, Chang, J.S, Han, S, Kurumbail, R, Thorarenson, A. | | Deposit date: | 2011-09-16 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Identification of Type-II Inhibitors Using Kinase Structures.
Chem.Biol.Drug Des., 80, 2012
|
|
3TUC
 
 | | Crystal structure of SYK kinase domain with 1-benzyl-N-(5-(6,7-dimethoxyquinolin-4-yloxy)pyridin-2-yl)-2-oxo-1,2-dihydropyridine-3-carboxamide | | Descriptor: | 1-benzyl-N-{5-[(6,7-dimethoxyquinolin-4-yl)oxy]pyridin-2-yl}-2-oxo-1,2-dihydropyridine-3-carboxamide, Tyrosine-protein kinase SYK | | Authors: | Lovering, F, McDonald, J, Whitlock, G, Glossop, P, Phillips, C, Sabnis, Y, Ryan, M, Fitz, L, Lee, J, Chang, J.S, Han, S, Kurumbail, R, Thorarenson, A. | | Deposit date: | 2011-09-16 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Type-II Inhibitors Using Kinase Structures.
Chem.Biol.Drug Des., 80, 2012
|
|
