7W1Y
 
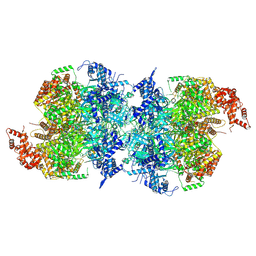 | | Human MCM double hexamer bound to natural DNA duplex (polyAT/polyTA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (49-MER), ... | | Authors: | Li, J, Dong, J, Dang, S, Zhai, Y. | | Deposit date: | 2021-11-21 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | The human pre-replication complex is an open complex.
Cell, 186, 2023
|
|
7V3U
 
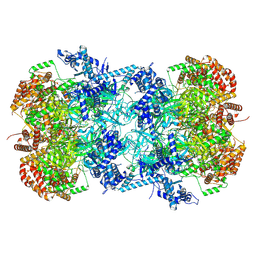 | | Cryo-EM structure of MCM double hexamer with structured Mcm4-NSD | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, ... | | Authors: | Cheng, J, Li, N, Huo, Y, Dang, S, Tye, B, Gao, N, Zhai, Y. | | Deposit date: | 2021-08-11 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Insight into the MCM double hexamer activation by Dbf4-Cdc7 kinase.
Nat Commun, 13, 2022
|
|
7V3V
 
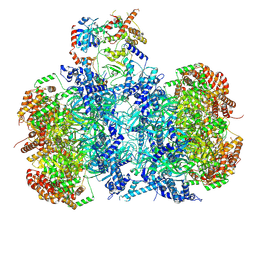 | | Cryo-EM structure of MCM double hexamer bound with DDK in State I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 7, DDK kinase regulatory subunit DBF4, ... | | Authors: | Cheng, J, Li, N, Huo, Y, Dang, S, Tye, B, Gao, N, Zhai, Y. | | Deposit date: | 2021-08-11 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Insight into the MCM double hexamer activation by Dbf4-Cdc7 kinase.
Nat Commun, 13, 2022
|
|
8GPN
 
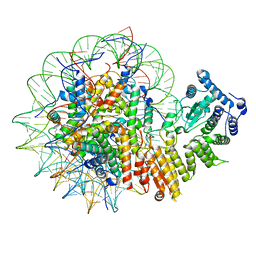 | | Human menin in complex with H3K79Me2 nucleosome | | Descriptor: | DNA (177-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Lin, J, Yu, D, Lam, W.H, Dang, S, Zhai, Y, Li, X.D. | | Deposit date: | 2022-08-26 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Menin "reads" H3K79me2 mark in a nucleosomal context.
Science, 379, 2023
|
|
3LRB
 
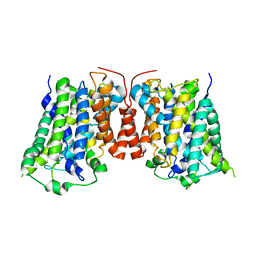 | | Structure of E. coli AdiC | | Descriptor: | Arginine/agmatine antiporter | | Authors: | Gao, X, Lu, F, Zhou, L, Shi, Y. | | Deposit date: | 2010-02-10 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Structure and mechanism of an amino acid antiporter
Science, 324, 2009
|
|
3LRC
 
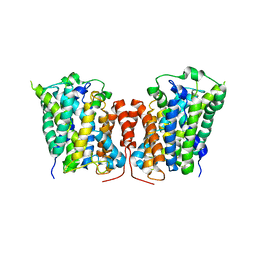 | | Structure of E. coli AdiC (P1) | | Descriptor: | Arginine/agmatine antiporter | | Authors: | Gao, X, Lu, F, Zhou, L, Shi, Y. | | Deposit date: | 2010-02-11 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.004 Å) | | Cite: | Structure and mechanism of an amino acid antiporter
Science, 324, 2009
|
|
3QE7
 
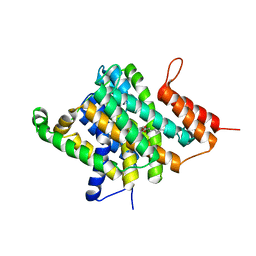 | | Crystal Structure of Uracil Transporter--UraA | | Descriptor: | URACIL, Uracil permease, nonyl beta-D-glucopyranoside | | Authors: | Lu, F.R, Li, S, Yan, N. | | Deposit date: | 2011-01-20 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.781 Å) | | Cite: | Structure and mechanism of the uracil transporter UraA
Nature, 472, 2011
|
|
7XG7
 
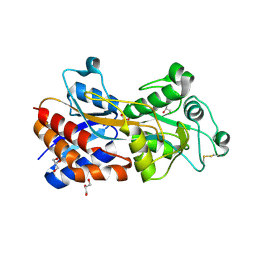 | | Crystal structure of PstS protein from cyanophage Syn19 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Phosphate transporter subunit | | Authors: | Cai, K, Jiang, Y.L. | | Deposit date: | 2022-04-03 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural characterization of the cyanophage-encoded phosphate-binding protein: implications for enhanced phosphate uptake of infected cyanobacteria
Environ.Microbiol., 24, 2022
|
|
7XG8
 
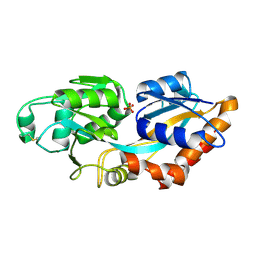 | | Crystal structure of PstS protein from cyanophage P-SSM2 | | Descriptor: | ABC transporter, substrate binding protein, phosphate, ... | | Authors: | Cai, K, Jiang, Y.L. | | Deposit date: | 2022-04-03 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Biochemical and structural characterization of the cyanophage-encoded phosphate-binding protein: implications for enhanced phosphate uptake of infected cyanobacteria
Environ.Microbiol., 24, 2022
|
|
8DW9
 
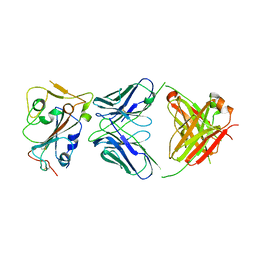 | |
8DXS
 
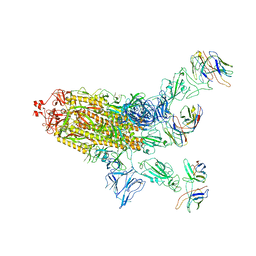 | |
8DWA
 
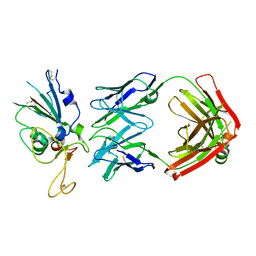 | |
7W8G
 
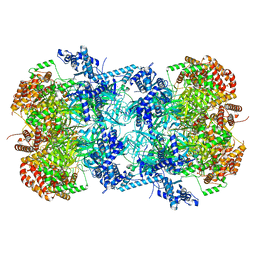 | | Cryo-EM structure of MCM double hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, ... | | Authors: | Cheng, J, Li, N, Tye, B, Zhai, Y, Gao, N. | | Deposit date: | 2021-12-07 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural Insight into the MCM double hexamer activation by Dbf4-Cdc7 kinase.
Nat Commun, 13, 2022
|
|
8HGO
 
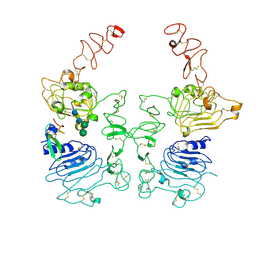 | | The EGF-bound EGFR/HER2 ectodomain complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor, ... | | Authors: | Zhang, Z, Bai, X. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structure and dynamics of the EGFR/HER2 heterodimer.
Cell Discov, 9, 2023
|
|
8HGP
 
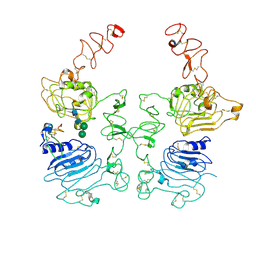 | | The EREG-bound EGFR/HER2 ectodomain complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Zhang, Z, Bai, X. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.53 Å) | | Cite: | Structure and dynamics of the EGFR/HER2 heterodimer.
Cell Discov, 9, 2023
|
|
8HGS
 
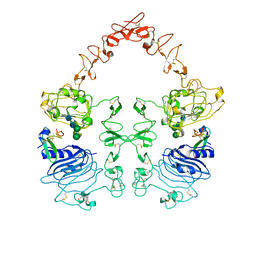 | | The EGF-bound EGFR ectodomain homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, Pro-epidermal growth factor, ... | | Authors: | Zhang, Z, Bai, X. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structure and dynamics of the EGFR/HER2 heterodimer.
Cell Discov, 9, 2023
|
|
8XGC
 
 | | Structure of yeast replisome associated with FACT and histone hexamer, Composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, Chromosome segregation in meiosis protein 3, ... | | Authors: | Li, N, Gao, Y, Yu, D, Gao, N, Zhai, Y. | | Deposit date: | 2023-12-15 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Parental histone transfer caught at the replication fork.
Nature, 627, 2024
|
|
4M6O
 
 | | Crystal structure of anti-NGF antibody CNTO7309 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CNTO7309 heavy chain, CNTO7309 light chain, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-08-09 | | Release date: | 2014-03-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
4M7K
 
 | | Crystal structure of anti-tissue factor antibody 10H10 | | Descriptor: | 10H10 heavy chain, 10H10 light chain, ACETATE ION, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-08-12 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
4M6M
 
 | | Crystal structure of anti-IL-23 antibody CNTO1959 at pH 9.5 | | Descriptor: | CNTO1959 heavy chain, CNTO1959 light chain, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L. | | Deposit date: | 2013-08-09 | | Release date: | 2014-03-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
4MAU
 
 | | Crystal structure of anti-ST2L antibody C2244 | | Descriptor: | C2244 heavy chain, C2244 light chain, FORMIC ACID, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-08-16 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
4KMT
 
 | | Crystal structure of human germline antibody 5-51/O12 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-05-08 | | Release date: | 2014-03-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
4KQ4
 
 | | Crystal structure of Anti-IL-17A antibody CNTO7357 | | Descriptor: | CNTO7357 heavy chain, CNTO7357 light chain, NICKEL (II) ION | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-05-14 | | Release date: | 2014-03-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
