5A63
 
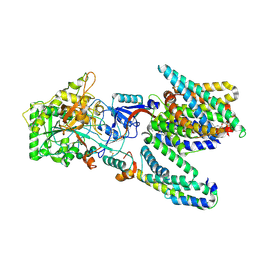 | | Cryo-EM structure of the human gamma-secretase complex at 3.4 angstrom resolution. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bai, X, Yan, C, Yang, G, Lu, P, Ma, D, Sun, L, Zhou, R, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An Atomic Structure of Human Gamma-Secretase
Nature, 525, 2015
|
|
3Q0K
 
 | | Crystal structure of Human PACSIN 2 F-BAR | | Descriptor: | CALCIUM ION, Protein kinase C and casein kinase substrate in neurons protein 2 | | Authors: | Bai, X, Meng, G, Zheng, X. | | Deposit date: | 2010-12-15 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human PACSIN 2 F-BAR domain
To be Published
|
|
3SYV
 
 | | Crystal structure of mPACSIN 3 F-BAR domain mutant | | Descriptor: | Protein kinase C and casein kinase II substrate protein 3 | | Authors: | Bai, X. | | Deposit date: | 2011-07-18 | | Release date: | 2012-05-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Rigidity of wedge loop in PACSIN 3 protein is a key factor in dictating diameters of tubules
J.Biol.Chem., 287, 2012
|
|
3Q84
 
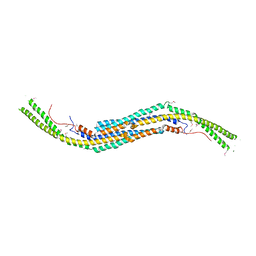 | | Crystal structure of human PACSIN 1 F-BAR domain | | Descriptor: | CALCIUM ION, Protein kinase C and casein kinase substrate in neurons protein 1 | | Authors: | Bai, X. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human PACSIN 1 F-BAR domain
To be Published
|
|
2OT5
 
 | |
7FJG
 
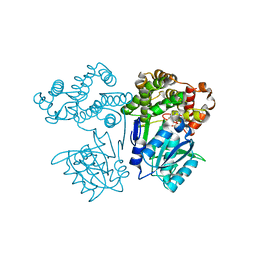 | | Crystal structure of butanol dehydrogenase A (YqdH) in complex with partial NADH from Fusobacterium nucleatum | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FE (III) ION, NADH-dependent butanol dehydrogenase A | | Authors: | Bai, X, Lan, J, Wang, L, Bu, T, Xu, Y. | | Deposit date: | 2021-08-03 | | Release date: | 2022-06-15 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of butanol dehydrogenase A (YqdH) in complex with partial NADH from Fusobacterium nucleatum
To Be Published
|
|
8I29
 
 | | Crystal structure of butanol dehydrogenase A (YqdH) in complex with NADH from Fusobacterium nucleatum | | Descriptor: | CHLORIDE ION, COBALT (II) ION, NADH-dependent butanol dehydrogenase A, ... | | Authors: | Bai, X, Lan, J, Bu, T.T, Wang, L.L, He, S.R, Zhang, J, Xu, Y.B. | | Deposit date: | 2023-01-14 | | Release date: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of butanol dehydrogenase A (YqdH) in complex with NADH from Fusobacterium nucleatum
To Be Published
|
|
4V92
 
 | | Kluyveromyces lactis 80S ribosome in complex with CrPV-IRES | | Descriptor: | 18S RRNA, ES1, ES10, ... | | Authors: | Fernandez, I.S, Bai, X, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-03-21 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Initiation of Translation by Cricket Paralysis Virus Ires Requires its Translocation in the Ribosome.
Cell(Cambridge,Mass.), 157, 2014
|
|
4V91
 
 | | Kluyveromyces lactis 80S ribosome in complex with CrPV-IRES | | Descriptor: | 25S RRNA, 5.8S RRNA, 5S RRNA, ... | | Authors: | Fernandez, I.S, Bai, X, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-03-21 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Initiation of Translation by Cricket Paralysis Virus Ires Requires its Translocation in the Ribosome.
Cell(Cambridge,Mass.), 157, 2014
|
|
4AYI
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 3 Wild type | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H, LIPOPROTEIN GNA1870 CCOMPND 7 | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-21 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
4AYD
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 1 R106A mutant | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-20 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules
Plos Pathog., 8, 2012
|
|
4AYN
 
 | | Structure of the C-terminal barrel of Neisseria meningitidis FHbp Variant 2 | | Descriptor: | FACTOR H-BINDING PROTEIN, SULFATE ION | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-21 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
4AYM
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 3 P106A mutant | | Descriptor: | COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-21 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
4AYE
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 1 E283AE304A mutant | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-20 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
6NT9
 
 | | Cryo-EM structure of the complex between human TBK1 and chicken STING | | Descriptor: | Serine/threonine-protein kinase TBK1, Stimulator of interferon genes protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of STING binding with and phosphorylation by TBK1.
Nature, 567, 2019
|
|
6VXK
 
 | | Cryo-EM Structure of the full-length A39R/PlexinC1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-C1, Semaphorin-like protein 139 | | Authors: | Kuo, Y.-C, Chen, H, Shang, G, Uchikawa, E, Tian, H, Bai, X, Zhang, X. | | Deposit date: | 2020-02-22 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of the PlexinC1/A39R complex reveals inter-domain interactions critical for ligand-induced activation.
Nat Commun, 11, 2020
|
|
8EQB
 
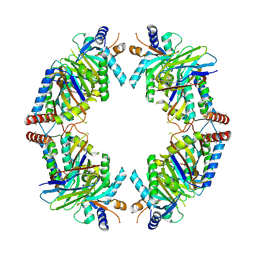 | | FAM46C/BCCIPalpha/Nanobody complex | | Descriptor: | Isoform 2 of BRCA2 and CDKN1A-interacting protein, Synthetic nanobody 1, Terminal nucleotidyltransferase 5C | | Authors: | Liu, S, Chen, H, Yin, Y, Bai, X, Zhang, X. | | Deposit date: | 2022-10-07 | | Release date: | 2023-03-15 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Inhibition of FAM46/TENT5 activity by BCCIP alpha adopting a unique fold.
Sci Adv, 9, 2023
|
|
3QE6
 
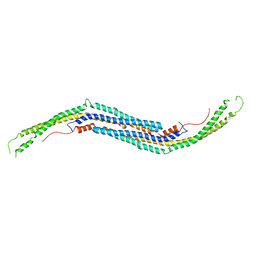 | | Mouse PACSIN 3 F-BAR domain structure | | Descriptor: | MAGNESIUM ION, Protein kinase C and casein kinase II substrate protein 3 | | Authors: | Meng, G, Bai, X, Zheng, X. | | Deposit date: | 2011-01-19 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Rigidity of wedge loop in PACSIN 3 protein is a key factor in dictating diameters of tubules
J.Biol.Chem., 287, 2012
|
|
3QNI
 
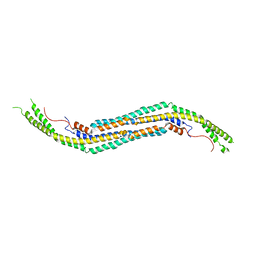 | |
3J8H
 
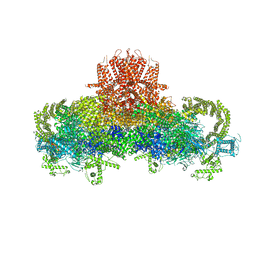 | | Structure of the rabbit ryanodine receptor RyR1 in complex with FKBP12 at 3.8 Angstrom resolution | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ZINC ION | | Authors: | Yan, Z, Bai, X, Yan, C, Wu, J, Scheres, S.H.W, Shi, Y, Yan, N. | | Deposit date: | 2014-10-26 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution.
Nature, 517, 2015
|
|
6NT5
 
 | | Cryo-EM structure of full-length human STING in the apo state | | Descriptor: | Stimulator of interferon protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
6NT6
 
 | | Cryo-EM structure of full-length chicken STING in the apo state | | Descriptor: | Stimulator of interferon genes protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
6NT7
 
 | | Cryo-EM structure of full-length chicken STING in the cGAMP-bound dimeric state | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
6NT8
 
 | | Cryo-EM structure of full-length chicken STING in the cGAMP-bound tetrameric state | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
8FLM
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP, NVS-STG2 and C53 | | Descriptor: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, ... | | Authors: | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
