2CKL
 
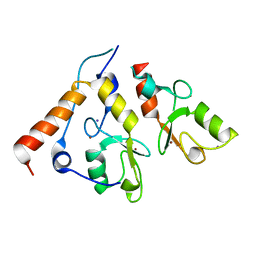 | | Ring1b-Bmi1 E3 catalytic domain structure | | Descriptor: | IODIDE ION, POLYCOMB GROUP RING FINGER PROTEIN 4, UBIQUITIN LIGASE PROTEIN RING2, ... | | Authors: | Buchwald, G, van der Stoop, P, Weichenrieder, O, Perrakis, A, van Lohuizen, M, Sixma, T.K. | | Deposit date: | 2006-04-20 | | Release date: | 2006-05-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and E3-Ligase Activity of the Ring-Ring Complex of Polycomb Proteins Bmi1 and Ring1B.
Embo J., 25, 2006
|
|
1TC3
 
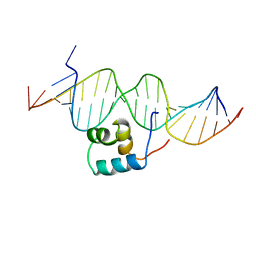 | | TRANSPOSASE TC3A1-65 FROM CAENORHABDITIS ELEGANS | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*GP*TP*CP*CP*TP*AP*TP*AP*GP*A P*AP*CP*TP*T)-3'), DNA (5'-D(*AP*GP*TP*TP*CP*TP*AP*TP*AP*GP*GP*AP*CP*CP*CP*CP*C P*CP*CP*T)-3'), PROTEIN (TC3 TRANSPOSASE) | | Authors: | Van Pouderoyen, G, Ketting, R.F, Perrakis, A, Plasterk, R.H.A, Sixma, T.K. | | Deposit date: | 1997-07-07 | | Release date: | 1997-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the specific DNA-binding domain of Tc3 transposase of C.elegans in complex with transposon DNA.
EMBO J., 16, 1997
|
|
1U9A
 
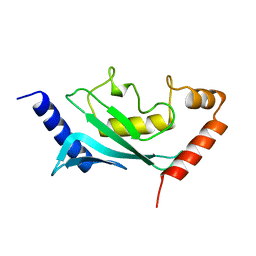 | | HUMAN UBIQUITIN-CONJUGATING ENZYME UBC9 | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME | | Authors: | Tong, H, Hateboer, G, Perrakis, A, Bernards, R, Sixma, T.K. | | Deposit date: | 1997-02-11 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of murine/human Ubc9 provides insight into the variability of the ubiquitin-conjugating system.
J.Biol.Chem., 272, 1997
|
|
1UGO
 
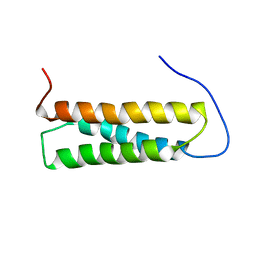 | | Solution structure of the first Murine BAG domain of Bcl2-associated athanogene 5 | | Descriptor: | Bcl2-associated athanogene 5 | | Authors: | Endoh, H, Hayashi, F, Seimiya, K, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-17 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The C-terminal BAG domain of BAG5 induces conformational changes of the Hsp70 nucleotide-binding domain for ADP-ATP exchange
Structure, 18, 2010
|
|
7ZXU
 
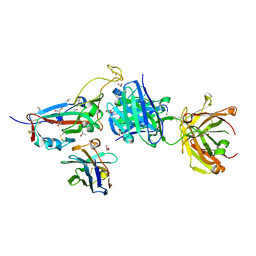 | | SARS-CoV-2 Omicron BA.4/5 RBD in complex with Beta-27 Fab and C1 nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-27 heavy chain, Beta-27 light chain, ... | | Authors: | Huo, J, Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-05-23 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum.
Cell, 185, 2022
|
|
1UK5
 
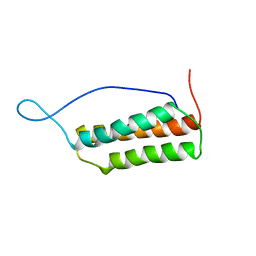 | | Solution structure of the Murine BAG domain of Bcl2-associated athanogene 3 | | Descriptor: | BAG-family molecular chaperone regulator-3 | | Authors: | Hatta, R, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-19 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The C-terminal BAG domain of BAG5 induces conformational changes of the Hsp70 nucleotide-binding domain for ADP-ATP exchange
Structure, 18, 2010
|
|
2D9D
 
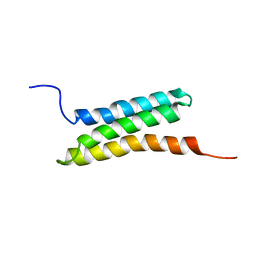 | | Solution structure of the BAG domain (275-350) of BAG-family molecular chaperone regulator-5 | | Descriptor: | BAG family molecular chaperone regulator 5 | | Authors: | Hatta, R, Hayashi, F, Yoshida, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-09 | | Release date: | 2006-12-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The C-terminal BAG domain of BAG5 induces conformational changes of the Hsp70 nucleotide-binding domain for ADP-ATP exchange
Structure, 18, 2010
|
|
5LO5
 
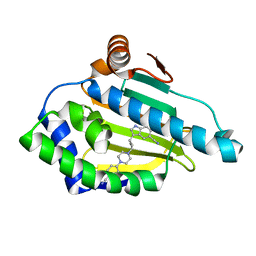 | | HSP90 WITH indole derivative | | Descriptor: | 3-[4-[4-(4-cyanophenyl)piperazin-1-yl]butyl]-6-oxidanyl-1~{H}-indole-5-carbonitrile, Heat shock protein HSP 90-alpha | | Authors: | Graedler, U, Amaral, M. | | Deposit date: | 2016-08-08 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
7Z6O
 
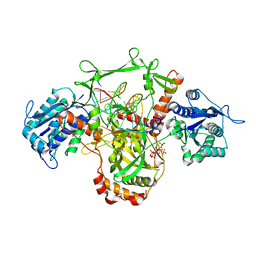 | | X-Ray studies of Ku70/80 reveal the binding site for IP6 | | Descriptor: | DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*AP*AP*AP*CP*TP*AP*AP*AP*AP*AP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Varela, P.F, Charbonnier, J.B. | | Deposit date: | 2022-03-14 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
7SFR
 
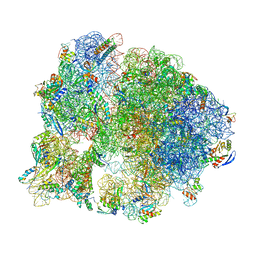 | | Unmethylated Mtb Ribosome 50S with SEQ-9 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Xing, Z, Cui, Z, Zhang, J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2021-10-04 | | Release date: | 2022-10-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Discovery of natural-product-derived sequanamycins as potent oral anti-tuberculosis agents.
Cell, 186, 2023
|
|
4F0I
 
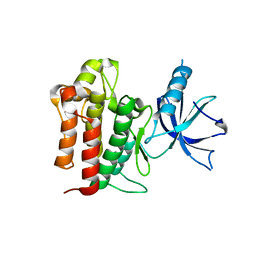 | | Crystal structure of apo TrkA | | Descriptor: | High affinity nerve growth factor receptor | | Authors: | Liu, J. | | Deposit date: | 2012-05-04 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | The Crystal Structures of TrkA and TrkB Suggest Key Regions for Achieving Selective Inhibition.
J.Mol.Biol., 423, 2012
|
|
4UMX
 
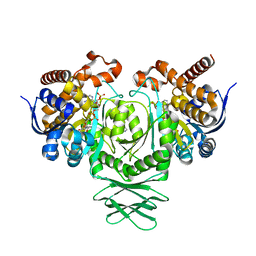 | | IDH1 R132H in complex with cpd 1 | | Descriptor: | 2,6-bis(1H-imidazol-1-ylmethyl)-4-(2,4,4-trimethylpentan-2-yl)phenol, GLYCEROL, ISOCITRATE DEHYDROGENASE [NADP] CYTOPLASMIC, ... | | Authors: | Mathieu, M, Marquette, J.P. | | Deposit date: | 2014-05-22 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Selective Inhibition of Mutant Isocitrate Dehydrogenase 1 (Idh1) Via Disruption of a Metal Binding Network by an Allosteric Small Molecule.
J.Biol.Chem., 290, 2015
|
|
4UMY
 
 | | IDH1 R132H in complex with cpd 1 | | Descriptor: | GLYCEROL, ISOCITRATE DEHYDROGENASE [NADP] CYTOPLASMIC, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | McLean, L, Zhang, Y, Mathieu, M. | | Deposit date: | 2014-05-22 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Selective Inhibition of Mutant Isocitrate Dehydrogenase 1 (Idh1) Via Disruption of a Metal Binding Network by an Allosteric Small Molecule.
J.Biol.Chem., 290, 2015
|
|
7KGB
 
 | |
5LO6
 
 | | HSP90 WITH indazole derivative | | Descriptor: | 3-(3,3-dimethylbutyl)-~{N}-methyl-~{N}-[4-(1-methylpiperidin-4-yl)phenyl]-6-oxidanyl-2~{H}-indazole-5-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Graedler, U, Amaral, M. | | Deposit date: | 2016-08-08 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
6RO0
 
 | | CRYSTAL STRUCTURE OF GENETICALLY DETOXIFIED PERTUSSIS TOXIN GDPT. | | Descriptor: | GLYCEROL, Islet-activating protein S2, Islet-activating protein S3, ... | | Authors: | Bertrand, T. | | Deposit date: | 2019-05-10 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Genetically detoxified pertussis toxin displays near identical structure to its wild-type and exhibits robust immunogenicity.
Commun Biol, 3, 2020
|
|
6I9J
 
 | | Human transforming growth factor beta2 in a tetragonal crystal form | | Descriptor: | Transforming growth factor beta-2 proprotein | | Authors: | Gomis-Ruth, F.X, Marino-Puertas, L, del Amo-Maestro, L, Goulas, T. | | Deposit date: | 2018-11-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recombinant production, purification, crystallization, and structure analysis of human transforming growth factor beta 2 in a new conformation.
Sci Rep, 9, 2019
|
|
7ZT6
 
 | |
7ZVT
 
 | | CryoEM structure of Ku heterodimer bound to DNA | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*TP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-17 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
4KV5
 
 | | scFv GC1009 in complex with TGF-beta1. | | Descriptor: | Single-chain variable fragment GC1009, Transforming growth factor beta-1 proprotein | | Authors: | Wei, R, Moulin, A.G, Mathieu, M. | | Deposit date: | 2013-05-22 | | Release date: | 2014-09-24 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of a pan-specific antagonist antibody complexed to different isoforms of TGF beta reveal structural plasticity of antibody-antigen interactions.
Protein Sci., 23, 2014
|
|
4KXZ
 
 | | crystal structure of tgfb2 in complex with GC2008. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, GC1008 Heavy Chain, ... | | Authors: | Mathieu, M, Moulin, A.G, Wei, R. | | Deposit date: | 2013-05-28 | | Release date: | 2014-09-24 | | Last modified: | 2014-12-03 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structures of a pan-specific antagonist antibody complexed to different isoforms of TGF beta reveal structural plasticity of antibody-antigen interactions.
Protein Sci., 23, 2014
|
|
5HNI
 
 | | CRYSTAL STRUCTURE OF CMET WT with compound 3 | | Descriptor: | Hepatocyte growth factor receptor, methyl (6-{[6-(4-fluorophenyl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1H-benzimidazol-2-yl)carbamate | | Authors: | Vallee, F, Houtmann, J. | | Deposit date: | 2016-01-18 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
5HO6
 
 | | CRYSTAL STRUCTURE OF CMET IN COMPLEX WITH CMPD. | | Descriptor: | 1-(6-{[6-(4-fluorophenyl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1,3-benzothiazol-2-yl)-3-[2-(morpholin-4-yl)ethyl]urea, Hepatocyte growth factor receptor | | Authors: | Vallee, F, Houtmann, J. | | Deposit date: | 2016-01-19 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
5HOR
 
 | | Crystal structure of c-Met-M1250T in complex with SAR125844. | | Descriptor: | 1-(6-{[6-(4-fluorophenyl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1,3-benzothiazol-2-yl)-3-[2-(morpholin-4-yl)ethyl]urea, Hepatocyte growth factor receptor | | Authors: | Vallee, F, Houtmann, J, Marquette, J.-P. | | Deposit date: | 2016-01-19 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
5HLW
 
 | | Crystal structure of c-Met mutant Y1230H in complex with compound 14 | | Descriptor: | 1-[2-(1-ethylpiperidin-4-yl)ethyl]-3-(6-{[6-(thiophen-2-yl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1,3-benzothiazol-2-yl)urea, CHLORIDE ION, Hepatocyte growth factor receptor | | Authors: | Vallee, F, Pouzieux, S, Marquette, J.P, Houtmann, J. | | Deposit date: | 2016-01-15 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
