5CEH
 
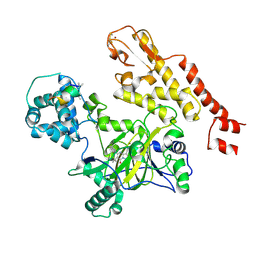 | | Structure of histone lysine demethylase KDM5A in complex with selective inhibitor | | Descriptor: | 7-oxo-5-phenyl-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile, Lysine-specific demethylase 5A, NICKEL (II) ION, ... | | Authors: | Kiefer, J.R, Vinogradova, M. | | Deposit date: | 2015-07-06 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells.
Nat.Chem.Biol., 12, 2016
|
|
6MJZ
 
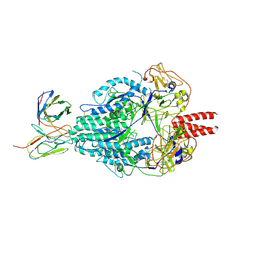 | | Cryo-EM structure of Human Parainfluenza Virus Type 3 (hPIV3) in complex with antibody PIA174 | | Descriptor: | Fusion glycoprotein F0, PIA174 Fab Heavy chain, PIA174 Fab Light chain | | Authors: | Acharya, P, Stewart-Jones, G, Carragher, B, Potter, C.S, Kwong, P.D. | | Deposit date: | 2018-09-24 | | Release date: | 2018-11-14 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure-based design of a quadrivalent fusion glycoprotein vaccine for human parainfluenza virus types 1-4.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8VPG
 
 | |
8VPI
 
 | |
8VPH
 
 | |
6WV6
 
 | | Human VKOR with phenindione | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Phenindione, Vitamin K epoxide reductase, ... | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WV5
 
 | | Human VKOR C43S mutant with vitamin K1 epoxide | | Descriptor: | (2R,3R)-2-hydroxy-3-methyl-2-[(2E,7S)-3,7,11,15-tetramethylhexadec-2-en-1-yl]-2,3-dihydronaphthalene-1,4-dione, Vitamin K epoxide reductase Cys43Ser mutant, termini restrained by green fluorescent protein | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WV3
 
 | | Human VKOR with warfarin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, S-WARFARIN, Vitamin K epoxide reductase, ... | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WVH
 
 | | Human VKOR with Brodifacoum | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Brodifacoum, Vitamin K epoxide reductase, ... | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WVB
 
 | | Takifugu rubripes VKOR-like with warfarin | | Descriptor: | S-WARFARIN, Vitamin K epoxide reductase-like protein, termini restrained by green fluorescent protein | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.872 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6JDQ
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA | | Descriptor: | CRISPR-associated endonuclease Cas9, sgRNA | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JE3
 
 | | Crystal structure of Nme2Cas9 in complex with sgRNA and target DNA (AGGCCC PAM) with 5 nt overhang | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, non-target DNA strand, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-03 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.931 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6KC7
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in seed-base paring state | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (5'-D(*AP*TP*AP*TP*GP*AP*TP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*AP*TP*CP*AP*TP*AP*TP*GP*TP*AP*AP*AP*GP*TP*T)-3'), ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-06-27 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
5WDM
 
 | |
6KC8
 
 | | Crystal structure of WT Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in post-cleavage state | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (5'-D(*AP*TP*AP*TP*GP*AP*TP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*AP*TP*CP*AP*TP*AP*TP*GP*TP*A)-3'), ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-06-27 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6L40
 
 | | Discovery of novel peptidomimetic boronate ClpP inhibitors with noncanonical enzyme mechanism as potent virulence blockers in vitro and in vivo | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, [(1S)-3-methyl-1-[[(2S)-3-phenyl-2-(pyrazin-2-ylcarbonylamino)propanoyl]amino]butyl]boronic acid | | Authors: | Luo, Y.F, Bao, R, Ju, Y, He, L.H. | | Deposit date: | 2019-10-15 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Discovery of Novel Peptidomimetic Boronate ClpP Inhibitors with Noncanonical Enzyme Mechanism as Potent Virulence Blockersin Vitroandin Vivo.
J.Med.Chem., 63, 2020
|
|
6JDV
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in catalytic state | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, MAGNESIUM ION, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JFU
 
 | | Crystal structure of Nme2Cas9 in complex with sgRNA and target DNA (AGGCCC PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, non-target strand, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-12 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
5TDL
 
 | |
6J1O
 
 | |
6J46
 
 | | LepI-SAH complex structure | | Descriptor: | O-methyltransferase lepI, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Qiu, S, Wei, C. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Deciphering the regulatory and catalytic mechanisms of an unusual SAM-dependent enzyme.
Signal Transduct Target Ther, 4, 2019
|
|
6JLC
 
 | |
6J24
 
 | | Crystal structure of a SAM-dependent methyltransferase LepI in complex with its substrate | | Descriptor: | (3~{S},4'~{R},4'~{a}~{S},6'~{R},8'~{a}~{S})-4',6'-dimethyl-5-phenyl-spiro[1~{H}-pyridine-3,5'-2,3,4,4~{a},6,8~{a}-hexahydro-1~{H}-naphthalene]-2,4-dione, O-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Qiu, S, Wei, C. | | Deposit date: | 2018-12-30 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Deciphering the regulatory and catalytic mechanisms of an unusual SAM-dependent enzyme.
Signal Transduct Target Ther, 4, 2019
|
|
5UTY
 
 | | Crystal Structure of a Stabilized DS-SOSIP.mut4 BG505 gp140 HIV-1 Env Trimer, Containing Mutations I201C-P433C (DS), L154M, N300M, N302M, T320L in Complex with Human Antibodies PGT122 and 35O22 at 4.1 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Fab heavy chain, ... | | Authors: | Xu, K, Chuang, G.-Y, Pancera, M, Kwong, P.D. | | Deposit date: | 2017-02-15 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.412 Å) | | Cite: | Structure-Based Design of a Soluble Prefusion-Closed HIV-1 Env Trimer with Reduced CD4 Affinity and Improved Immunogenicity.
J. Virol., 91, 2017
|
|
5VC8
 
 | | Crystal structure of the WHSC1 PWWP1 domain | | Descriptor: | DNA (5'-D(P*CP*TP*(DN))-3'), Histone-lysine N-methyltransferase NSD2, UNKNOWN ATOM OR ION, ... | | Authors: | Qin, S, Tempel, W, Dong, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Histone and DNA binding ability studies of the NSD subfamily of PWWP domains.
Biochem.Biophys.Res.Commun., 569, 2021
|
|
