8VM2
 
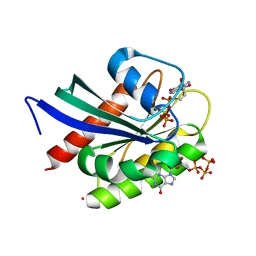 | | Crystal Structure of NRAS Q61K bound to GTP | | Descriptor: | GTPase NRas, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gebregiworgis, T, Chan, J.Y.L, Kuntz, D.A, Prive, G.G, Marshall, C.B, Ikura, M. | | Deposit date: | 2024-01-12 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of NRAS Q61K with a ligand-induced pocket near switch II.
Eur J Cell Biol, 103, 2024
|
|
5WI4
 
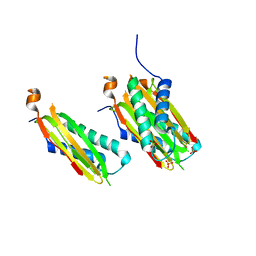 | | CRYSTAL STRUCTURE OF DYNLT1/TCTEX-1 IN COMPLEX WITH ARHGEF2 | | Descriptor: | Dynein light chain Tctex-type 1,Rho guanine nucleotide exchange factor 2, SULFATE ION | | Authors: | Balan, M, Ishiyama, N, Marshall, C.B, Ikura, M. | | Deposit date: | 2017-07-18 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | MARK3-mediated phosphorylation of ARHGEF2 couples microtubules to the actin cytoskeleton to establish cell polarity.
Sci Signal, 10, 2017
|
|
6W4F
 
 | | NMR-driven structure of KRAS4B-GDP homodimer on a lipid bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6W4E
 
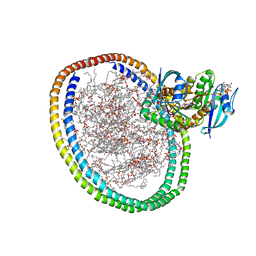 | | NMR-driven structure of KRAS4B-GTP homodimer on a lipid bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, ... | | Authors: | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
2K86
 
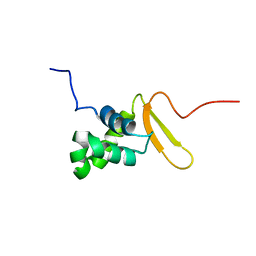 | | Solution Structure of FOXO3a Forkhead domain | | Descriptor: | Forkhead box protein O3 | | Authors: | Wang, F, Marshall, C.B, Li, G, Plevin, M.J, Ikura, M. | | Deposit date: | 2008-09-02 | | Release date: | 2008-10-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Biochemical and structural characterization of an intramolecular interaction in FOXO3a and its binding with p53.
J.Mol.Biol., 384, 2008
|
|
7RSE
 
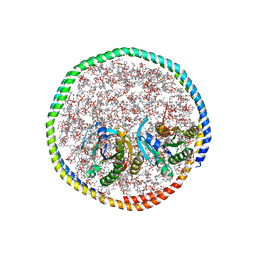 | | NMR-driven structure of the KRAS4B-G12D "alpha-beta" dimer on a lipid bilayer nanodisc | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Enomoto, M, Gebregiworgis, T, Gasmi-Seabrook, G.M, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Oncogenic KRAS G12D mutation promotes dimerization through a second, phosphatidylserine-dependent interface: a model for KRAS oligomerization.
Chem Sci, 12, 2021
|
|
7RSC
 
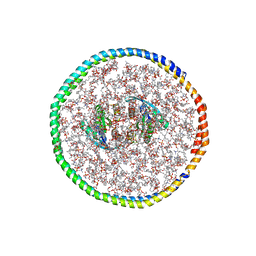 | | NMR-driven structure of the KRAS4B-G12D "alpha-alpha" dimer on a lipid bilayer nanodisc | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Enomoto, M, Gebregiworgis, T, Gasmi-Seabrook, G.M, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Oncogenic KRAS G12D mutation promotes dimerization through a second, phosphatidylserine-dependent interface: a model for KRAS oligomerization.
Chem Sci, 12, 2021
|
|
4O2L
 
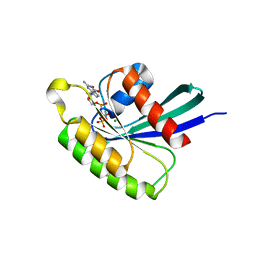 | | Structure of Mus musculus Rheb G63A mutant bound to GTP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GTP-binding protein Rheb, ... | | Authors: | Mazhab-Jafari, M.T, Marshall, C.B, Ho, J, Ishiyama, N, Stambolic, V, Ikura, M. | | Deposit date: | 2013-12-17 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-guided mutation of the conserved G3-box glycine in Rheb generates a constitutively activated regulator of mammalian target of rapamycin (mTOR).
J.Biol.Chem., 289, 2014
|
|
4O2R
 
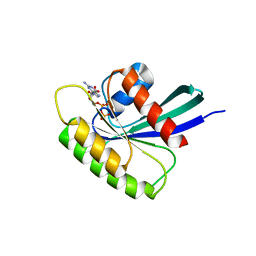 | | Structure of Mus musculus Rheb G63V mutant bound to GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Mazhab-Jafari, M.T, Marshall, C.B, Ho, J, Ishiyama, N, Stambolic, V, Ikura, M. | | Deposit date: | 2013-12-17 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-guided mutation of the conserved G3-box glycine in Rheb generates a constitutively activated regulator of mammalian target of rapamycin (mTOR).
J.Biol.Chem., 289, 2014
|
|
7SCX
 
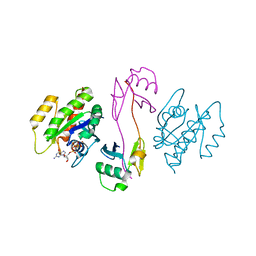 | | KRAS full-length G12V in complex with RGL1 Ras association domain | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | Authors: | Eves, B.J, Kuntz, D.A, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-09-29 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of RGL1 RAS-Association Domain in Complex with KRAS and the Oncogenic G12V Mutant.
J.Mol.Biol., 434, 2022
|
|
7SCW
 
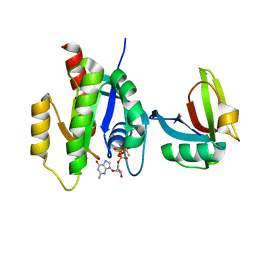 | | KRAS full length wild-type in complex with RGL1 Ras association domain | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | Authors: | Eves, B.J, Kuntz, D.A, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-09-29 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of RGL1 RAS-Association Domain in Complex with KRAS and the Oncogenic G12V Mutant.
J.Mol.Biol., 434, 2022
|
|
4O25
 
 | | Structure of Wild Type Mus musculus Rheb bound to GTP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Mazhab-Jafari, M.T, Marshall, C.B, Ho, J, Ishiyama, N, Stambolic, V, Ikura, M. | | Deposit date: | 2013-12-16 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-guided mutation of the conserved G3-box glycine in Rheb generates a constitutively activated regulator of mammalian target of rapamycin (mTOR).
J.Biol.Chem., 289, 2014
|
|
6PTS
 
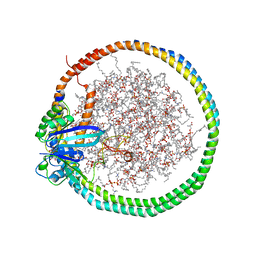 | | NMR data-driven model of KRas-GMPPNP:RBD-CRD complex tethered to a nanodisc (state A) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Fang, Z, Lee, K, Gasmi-Seabrook, G, Ikura, M, Marshall, C.B. | | Deposit date: | 2019-07-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multivalent assembly of KRAS with the RAS-binding and cysteine-rich domains of CRAF on the membrane.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PTW
 
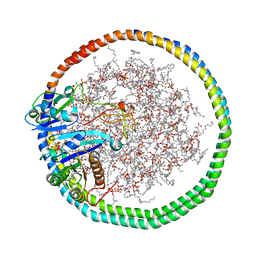 | | NMR data-driven model of KRas-GMPPNP:RBD-CRD complex tethered to a nanodisc (state B) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Fang, Z, Lee, K, Gasmi-Seabrook, G, Ikura, M, Marshall, C.B. | | Deposit date: | 2019-07-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multivalent assembly of KRAS with the RAS-binding and cysteine-rich domains of CRAF on the membrane.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2LQI
 
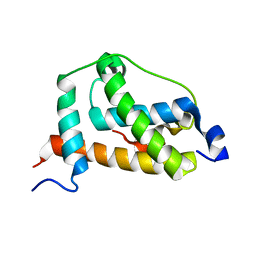 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2l3b conformation) | | Descriptor: | CREB-binding protein, Forkhead box O3 | | Authors: | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | Deposit date: | 2012-03-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LQH
 
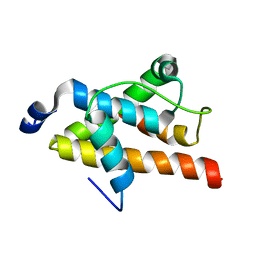 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2b3l conformation) | | Descriptor: | CREB-binding protein, Forkhead box O3 | | Authors: | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | Deposit date: | 2012-03-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6CC9
 
 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | Descriptor: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-06 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
6CCX
 
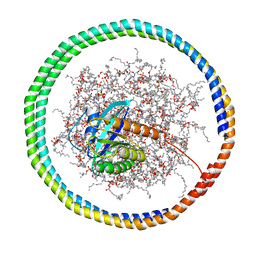 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | Descriptor: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
6CCH
 
 | | NMR data-driven model of GTPase KRas-GMPPNP tethered to a nanodisc (E3 state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
3SEA
 
 | | Structure of Rheb-Y35A mutant in GDP- and GMPPNP-bound forms | | Descriptor: | ACETATE ION, GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mazhab-Jafari, M.T, Marshall, C.B, Ishiyama, N, Vuk, S, Ikura, M. | | Deposit date: | 2011-06-10 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Autoinhibited Noncanonical Mechanism of GTP Hydrolysis by Rheb Maintains mTORC1 Homeostasis.
Structure, 20, 2012
|
|
6OS4
 
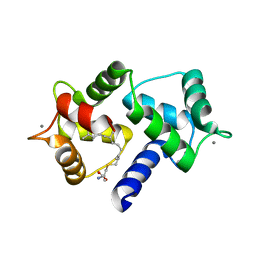 | | Calmodulin in complex with farnesyl cysteine methyl ester | | Descriptor: | CALCIUM ION, Calmodulin-1, s-farnesyl-l-cysteine methyl ester | | Authors: | Grant, B.M.M, Enomoto, M, Lee, K.Y, Back, S.I, Gebregiworgis, T, Ishiyama, N, Ikura, M, Marshall, C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Calmodulin disrupts plasma membrane localization of farnesylated KRAS4b by sequestering its lipid moiety.
Sci.Signal., 13, 2020
|
|
4KE2
 
 | | Crystal structure of the hyperactive Type I antifreeze from winter flounder | | Descriptor: | Type I hyperactive antifreeze protein | | Authors: | Sun, T, Lin, F.-H, Campbell, R.L, Allingham, J.S, Davies, P.L. | | Deposit date: | 2013-04-25 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An antifreeze protein folds with an interior network of more than 400 semi-clathrate waters.
Science, 343, 2014
|
|
2MSD
 
 | | NMR data-driven model of GTPase KRas-GNP tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MSE
 
 | | NMR data-driven model of GTPase KRas-GNP:ARafRBD complex tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MSC
 
 | | NMR data-driven model of GTPase KRas-GDP tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
