4JDV
 
 | | Crystal structure of germ-line precursor of NIH45-46 Fab | | Descriptor: | Fab heavy chain, Fab light chain, GLYCEROL, ... | | Authors: | Diskin, R, Scharf, L, Bjorkman, P.J. | | Deposit date: | 2013-02-25 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JKP
 
 | |
7MXE
 
 | | Ab1245 Fab in complex with BG505 SOSIP.664 and 8ANC195 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 8ANC195 G52K5 Fab heavy chain, ... | | Authors: | Abernathy, M.E, Bjorkman, P.J. | | Deposit date: | 2021-05-19 | | Release date: | 2021-10-27 | | Last modified: | 2021-11-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Antibody elicited by HIV-1 immunogen vaccination in macaques displaces Env fusion peptide and destroys a neutralizing epitope.
Npj Vaccines, 6, 2021
|
|
5THR
 
 | | Cryo-EM structure of a BG505 Env-sCD4-17b-8ANC195 complex | | Descriptor: | 17b Fab VH domain, 17b Fab VL domain, 8ANC195 G52K5 VH domain, ... | | Authors: | Wang, H, Bjorkman, P.J. | | Deposit date: | 2016-09-30 | | Release date: | 2016-11-16 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T3X
 
 | |
5UEM
 
 | | Crystal structure of 354NC37 Fab in complex with HIV-1 clade AE strain 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 354NC37 Fab Heavy Chain, ... | | Authors: | Sievers, S.A, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2017-01-02 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller.
Sci Transl Med, 9, 2017
|
|
5T3Z
 
 | |
5UEL
 
 | | Crystal structure of 354NC102 Fab | | Descriptor: | 354NC102 Fab Heavy Chain, 354NC102 Fab Light Chain, SULFATE ION | | Authors: | Sievers, S.A, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2017-01-02 | | Release date: | 2017-02-01 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller.
Sci Transl Med, 9, 2017
|
|
5UD9
 
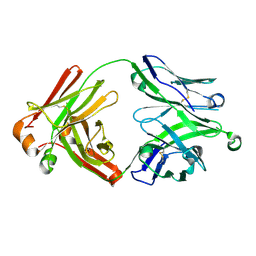 | | Crystal structure of 354BG18 Fab | | Descriptor: | Fab heavy chain, Light chain | | Authors: | Scharf, L, Bjorkman, P.J. | | Deposit date: | 2016-12-23 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller.
Sci Transl Med, 9, 2017
|
|
6UDK
 
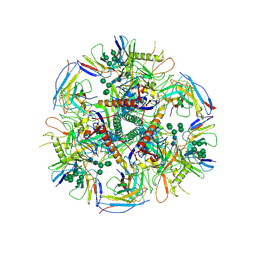 | | HIV-1 bNAb 1-55 in complex with modified BG505 SOSIP-based immunogen RC1 and 10-1074 | | Descriptor: | 1-55 Fab Heavy Chain, 1-55 Fab Light Chain, 10-1074 Fab Heavy Chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2019-09-19 | | Release date: | 2020-01-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Restriction of HIV-1 Escape by a Highly Broad and Potent Neutralizing Antibody.
Cell, 180, 2020
|
|
6UDJ
 
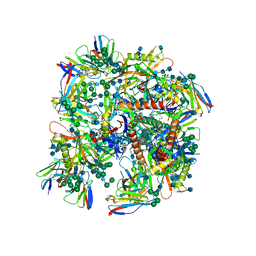 | | HIV-1 bNAb 1-18 in complex with BG505 SOSIP.664 and 10-1074 | | Descriptor: | 1-18 Fab Heavy Chain, 1-18 Fab Light Chain, 10-1074 Fab Heavy Chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2019-09-19 | | Release date: | 2020-01-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Restriction of HIV-1 Escape by a Highly Broad and Potent Neutralizing Antibody.
Cell, 180, 2020
|
|
6U0N
 
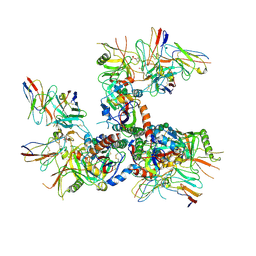 | |
6U0L
 
 | |
6URH
 
 | |
6UTA
 
 | | Crystal structure of Z004 iGL Fab in complex with ZIKV EDIII | | Descriptor: | Env, Z004 iGL Fab heavy chain, Z004 iGL Fab light chain | | Authors: | Esswein, S.R, Gristick, H.B, Keeffe, J.R, Bjorkman, P.J. | | Deposit date: | 2019-10-29 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for Zika envelope domain III recognition by a germline version of a recurrent neutralizing antibody.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UTE
 
 | | Crystal structure of Z032 Fab in complex with WNV EDIII | | Descriptor: | Envelope domain III, GLYCEROL, Z032 Fab heavy chain, ... | | Authors: | Esswein, S.R, Gristick, H.B, Keeffe, J.R, Bjorkman, P.J. | | Deposit date: | 2019-10-29 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for Zika envelope domain III recognition by a germline version of a recurrent neutralizing antibody.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4P9H
 
 | |
3S3Z
 
 | |
3S3Y
 
 | |
4Q6Y
 
 | | Crystal structure of a chemoenzymatic glycoengineered disialylated Fc (di-sFc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ahmed, A.A, Giddens, J, Pincetic, A, Lomino, J.V, Ravetch, J.V, Wang, L.X, Bjorkman, P.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural characterization of anti-inflammatory immunoglobulin g fc proteins.
J.Mol.Biol., 426, 2014
|
|
4Q74
 
 | | F241A Fc | | Descriptor: | Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ahmed, A.A, Giddens, J, Pincetic, A, Lomino, J.V, Ravetch, J.V, Wang, L.X, Bjorkman, P.J. | | Deposit date: | 2014-04-24 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural characterization of anti-inflammatory immunoglobulin g fc proteins.
J.Mol.Biol., 426, 2014
|
|
4P9M
 
 | | Crystal structure of 8ANC195 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8ANC195 Fab heavy chain, 8ANC195 light chain | | Authors: | Scharf, L, Bjorkman, P.J. | | Deposit date: | 2014-04-04 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Antibody 8ANC195 Reveals a Site of Broad Vulnerability on the HIV-1 Envelope Spike.
Cell Rep, 7, 2014
|
|
4Q7D
 
 | | Wild type Fc (wtFc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region | | Authors: | Ahmed, A.A, Giddens, J, Pincetic, A, Lomino, J.V, Ravetch, J.V, Wang, L.X, Bjorkman, P.J. | | Deposit date: | 2014-04-24 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of anti-inflammatory immunoglobulin g fc proteins.
J.Mol.Biol., 426, 2014
|
|
3RPI
 
 | | Crystal Structure of Fab from 3BNC60, Highly Potent anti-HIV Antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain from highly potent anti-HIV neutralizing antibody, Light chain from highly potent anti-HIV neutralizing antibody | | Authors: | Diskin, R, Bjorkman, P.J. | | Deposit date: | 2011-04-26 | | Release date: | 2011-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding.
Science, 333, 2011
|
|
3U7W
 
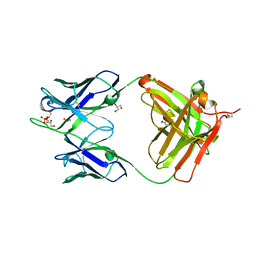 | | Crystal structure of NIH45-46 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Heavy chain, ... | | Authors: | Diskin, R, Bjorkman, P.J. | | Deposit date: | 2011-10-14 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Increasing the Potency and Breadth of an HIV Antibody by Using Structure-Based Rational Design.
Science, 334, 2011
|
|
