8Q84
 
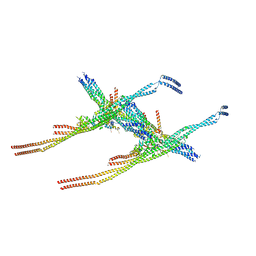 | |
8QAU
 
 | | Outer kinetochore Ndc80-Dam1 alpha/beta-tubulin complex | | Descriptor: | DASH complex subunit DAM1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Muir, K.W, Barford, D. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Mechanism of outer kinetochore assembly on microtubules and its regulation by mitotic error correction
Biorxiv, 2023
|
|
7DET
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Wang, Y, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
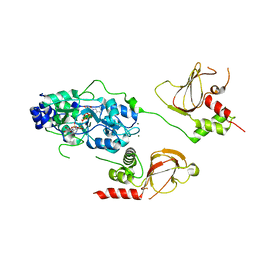
6N7E
 
 | |
5TWM
 
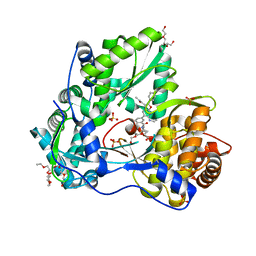 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS GENOTYPE 2A STRAIN JFH1 L30S NS5B RNA-DEPENDENT RNA POLYMERASE IN COMPLEX WITH 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide, NS5B RNA-dependent RNA POLYMERASE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The discovery of a pan-genotypic, primer grip inhibitor of HCV NS5B polymerase.
Medchemcomm, 8, 2017
|
|
6NM0
 
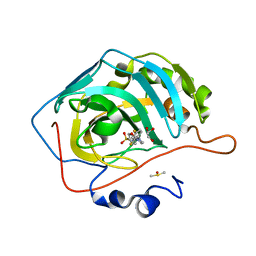 | | Selective inhibition of carbonic anhydrase IX activity, using compound SLC-149, displays limited anticancer effects in breast cancer cell lines | | Descriptor: | 4-[3-(2,4-difluorophenyl)-2-oxo-2,3-dihydro-1H-imidazol-1-yl]benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Singh, S, McKenna, R. | | Deposit date: | 2019-01-10 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Inhibition of Carbonic Anhydrase Using SLC-149: Support for a Noncatalytic Function of CAIX in Breast Cancer.
J.Med.Chem., 64, 2021
|
|
6MN6
 
 | |
6NLV
 
 | | Selective inhibition of carbonic anhydrase IX activity, using compound SLC-149, displays limited anticancer effects in breast cancer cell lines | | Descriptor: | 4-[3-(2,4-difluorophenyl)-2-oxo-2,3-dihydro-1H-imidazol-1-yl]benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Singh, S, McKenna, R. | | Deposit date: | 2019-01-09 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Inhibition of Carbonic Anhydrase Using SLC-149: Support for a Noncatalytic Function of CAIX in Breast Cancer.
J.Med.Chem., 64, 2021
|
|
7DEO
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Spike protein S1, ... | | Authors: | Fu, D, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
3E7J
 
 | | HeparinaseII H202A/Y257A double mutant complexed with a heparan sulfate tetrasaccharide substrate | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Heparinase II protein, ... | | Authors: | Shaya, D, Cygler, M. | | Deposit date: | 2008-08-18 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalytic mechanism of heparinase II investigated by site-directed mutagenesis and the crystal structure with its substrate.
J.Biol.Chem., 285, 2010
|
|
8II0
 
 | | FACTOR INHIBITING HIF-1 ALPHA in complex with (5-(3-(3-chlorophenyl)isoxazol-5-yl)-3-hydroxypicolinoyl)glycine | | Descriptor: | 2-[[5-[3-(3-chlorophenyl)-1,2-oxazol-5-yl]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Corner, T, Zhang, X, Schofield, C.J. | | Deposit date: | 2023-02-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | A Small-Molecule Inhibitor of Factor Inhibiting HIF Binding to a Tyrosine-Flip Pocket for the Treatment of Obesity.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8IHZ
 
 | | FACTOR INHIBITING HIF-1 ALPHA in complex with (5-(1-(3-(4-chlorophenyl)propyl)-1H-1,2,3-triazol-4-yl)-3-hydroxypicolinoyl)glycine | | Descriptor: | 2-[[5-[1-[3-(4-chlorophenyl)propyl]-1,2,3-triazol-4-yl]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Nakashima, Y, Corner, T, Zhang, X, Schofield, C.J. | | Deposit date: | 2023-02-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A Small-Molecule Inhibitor of Factor Inhibiting HIF Binding to a Tyrosine-Flip Pocket for the Treatment of Obesity.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
7EQ2
 
 | | Crystal structure of GDP-bound Rab1a-T75D | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Cao, Y.L, Gu, D.D, Gao, S. | | Deposit date: | 2021-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55090284 Å) | | Cite: | Aurora kinase A-mediated phosphorylation triggers structural alteration of Rab1A to enhance ER complexity during mitosis
Nat.Struct.Mol.Biol., 2024
|
|
5W99
 
 | | Pyridine synthase, PbtD, from GE2270 biosynthesis bound to TSP | | Descriptor: | 2,2'-(6-(2'-(aminomethyl)-[2,4'-bithiazol]-4-yl)pyridine-2,5-diyl)bis(thiazole-4-carboxylic acid), PbtD, SULFATE ION | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2017-06-22 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights into enzymatic [4+2] aza-cycloaddition in thiopeptide antibiotic biosynthesis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5Y81
 
 | | NuA4 TEEAA sub-complex | | Descriptor: | Actin, Actin-related protein 4, Chromatin modification-related protein EAF1, ... | | Authors: | Wang, X, Cai, G. | | Deposit date: | 2017-08-18 | | Release date: | 2018-04-18 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Architecture of the Saccharomyces cerevisiae NuA4/TIP60 complex
Nat Commun, 9, 2018
|
|
5UD8
 
 | | Crystal Structure of Mutant Ig-like Domain | | Descriptor: | Triggering receptor expressed on myeloid cells 2 | | Authors: | Sudom, A, Min, X, Wang, Z. | | Deposit date: | 2016-12-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis for the loss-of-function effects of the Alzheimer's disease-associated R47H variant of the immune receptor TREM2.
J. Biol. Chem., 293, 2018
|
|
5UD7
 
 | | Crystal Structure of Wild-Type Ig-like Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IODIDE ION, SULFATE ION, ... | | Authors: | Sudom, A, Min, X, Wang, Z. | | Deposit date: | 2016-12-23 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.20002246 Å) | | Cite: | Molecular basis for the loss-of-function effects of the Alzheimer's disease-associated R47H variant of the immune receptor TREM2.
J. Biol. Chem., 293, 2018
|
|
5WA4
 
 | |
7D3Y
 
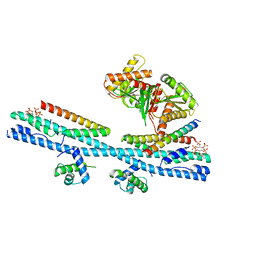 | | Crystal structure of the osPHR2-osSPX2 complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Protein PHOSPHATE STARVATION RESPONSE 2, SPX domain-containing protein 2,Isoform 1 of Core histone macro-H2A.1 | | Authors: | Zhang, Q.X, Guan, Z.Y, Zuo, J.Q, Zhang, Z.F, Liu, Z. | | Deposit date: | 2020-09-21 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Mechanistic insights into the regulation of plant phosphate homeostasis by the rice SPX2 - PHR2 complex.
Nat Commun, 13, 2022
|
|
8K3C
 
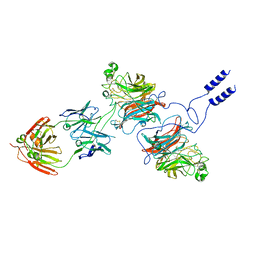 | | Nipah virus Attachment glycoprotein with 41-6 antibody fragment | | Descriptor: | Glycoprotein G, Heavy chain of 41-6 Fab fragments, Light chain of 41-6 Fab fragment | | Authors: | Sun, M.M. | | Deposit date: | 2023-07-15 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Potent human neutralizing antibodies against Nipah virus derived from two ancestral antibody heavy chains.
Nat Commun, 15, 2024
|
|
5W98
 
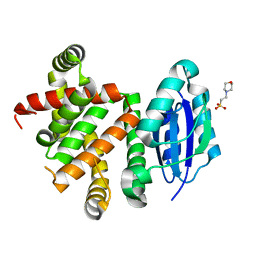 | | Pyridine synthase, PbtD, from GE2270 biosynthesis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PbtD | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2017-06-22 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structural insights into enzymatic [4+2] aza-cycloaddition in thiopeptide antibiotic biosynthesis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VMT
 
 | |
5WA3
 
 | |
3E80
 
 | |
5VQM
 
 | | Clostridium difficile TcdB-GTD bound to PA41 Fab | | Descriptor: | PA41 FAB HEAVY CHAIN, PA41 FAB LIGHT CHAIN, Toxin B | | Authors: | Kroh, H.K, Spiller, B.W, Lacy, D.B. | | Deposit date: | 2017-05-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A neutralizing antibody that blocks delivery of the enzymatic cargo of Clostridium difficile toxin TcdB into host cells.
J. Biol. Chem., 293, 2018
|
|
