6K3Z
 
 | | Crystal structure of dCas9 in complex with sgRNA and DNA (TGA PAM) | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (28-MER), DNA (5'-D(*AP*AP*AP*TP*GP*AP*TP*AP*TP*TP*G)-3'), ... | | Authors: | Chen, W, Zhang, H, Zhang, Y, Wang, Y, Gan, J, Ji, Q. | | Deposit date: | 2019-05-22 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular basis for the PAM expansion and fidelity enhancement of an evolved Cas9 nuclease.
Plos Biol., 17, 2019
|
|
6K4P
 
 | | Crystal structure of xCas9 in complex with sgRNA and DNA (TGG PAM) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (28-MER), PHOSPHATE ION, ... | | Authors: | Chen, W, Zhang, H, Zhang, Y, Wang, Y, Gan, J, Ji, Q. | | Deposit date: | 2019-05-25 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular basis for the PAM expansion and fidelity enhancement of an evolved Cas9 nuclease.
Plos Biol., 17, 2019
|
|
6K4U
 
 | | Crystal structure of xCas9 in complex with sgRNA and DNA (TGA PAM) | | Descriptor: | CRISPR-associated endonuclease Cas9, non-target DNA, sgRNA, ... | | Authors: | Chen, W, Zhang, H, Zhang, Y, Wang, Y, Gan, J, Ji, Q. | | Deposit date: | 2019-05-27 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular basis for the PAM expansion and fidelity enhancement of an evolved Cas9 nuclease.
Plos Biol., 17, 2019
|
|
2FZF
 
 | | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus | | Descriptor: | hypothetical protein | | Authors: | Fu, Z.-Q, Liu, Z.-J, Lee, D, Kelley, L, Chen, L, Tempel, W, Shah, N, Horanyi, P, Lee, H.S, Habel, J, Dillard, B.D, Nguyen, D, Chang, S.-H, Zhang, H, Chang, J, Sugar, F.J, Poole, F.L, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-02-09 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus
To be published
|
|
6K4Q
 
 | | Crystal structure of xCas9 in complex with sgRNA and DNA (CGG PAM) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (28-MER), DNA (5'-D(*AP*AP*AP*CP*GP*GP*TP*AP*TP*TP*G)-3'), ... | | Authors: | Chen, W, Zhang, H, Zhang, Y, Wang, Y, Gan, J, Ji, Q. | | Deposit date: | 2019-05-26 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for the PAM expansion and fidelity enhancement of an evolved Cas9 nuclease.
Plos Biol., 17, 2019
|
|
6K57
 
 | | Crystal structure of dCas9 in complex with sgRNA and DNA (CGA PAM) | | Descriptor: | CRISPR-associated endonuclease Cas9, non-target DNA, sgRNA, ... | | Authors: | Chen, W, Zhang, H, Zhang, Y, Wang, Y, Gan, J, Ji, Q. | | Deposit date: | 2019-05-28 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Molecular basis for the PAM expansion and fidelity enhancement of an evolved Cas9 nuclease.
Plos Biol., 17, 2019
|
|
6K4S
 
 | | Crystal structure of xCas9 in complex with sgRNA and DNA (TGC PAM) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, non-targeted DNA, sgRNA, ... | | Authors: | Chen, W, Zhang, H, Zhang, Y, Wang, Y, Gan, J, Ji, Q. | | Deposit date: | 2019-05-26 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Molecular basis for the PAM expansion and fidelity enhancement of an evolved Cas9 nuclease.
Plos Biol., 17, 2019
|
|
2HBX
 
 | | Crystal Structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde-Decarboxylase (ACMSD) | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | Authors: | Martynowski, D, Eyobo, Y, Li, T, Yang, K, Liu, A, Zhang, H. | | Deposit date: | 2006-06-14 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of alpha-Amino-beta-carboxymuconate-epsilon-semialdehyde Decarboxylase: Insight into the Active Site and Catalytic Mechanism of a Novel Decarboxylation Reaction.
Biochemistry, 45, 2006
|
|
2E76
 
 | | Crystal Structure of the Cytochrome b6f Complex with tridecyl-stigmatellin (TDS) from M.laminosus | | Descriptor: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 8-HYDROXY-5,7-DIMETHOXY-3-METHYL-2-TRIDECYL-4H-CHROMEN-4-ONE, ... | | Authors: | Cramer, W.A, Yamashita, E, Zhang, H. | | Deposit date: | 2007-01-05 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Structure of the Cytochrome b(6)f Complex: Quinone Analogue Inhibitors as Ligands of Heme c(n)
J.Mol.Biol., 370, 2007
|
|
2HBV
 
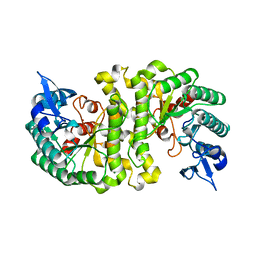 | | Crystal Structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde-Decarboxylase (ACMSD) | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, MAGNESIUM ION, ZINC ION | | Authors: | Martynowski, D, Eyobo, Y, Li, T, Yang, K, Liu, A, Zhang, H. | | Deposit date: | 2006-06-14 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of alpha-Amino-beta-carboxymuconate-epsilon-semialdehyde Decarboxylase: Insight into the Active Site and Catalytic Mechanism of a Novel Decarboxylation Reaction.
Biochemistry, 45, 2006
|
|
2I7T
 
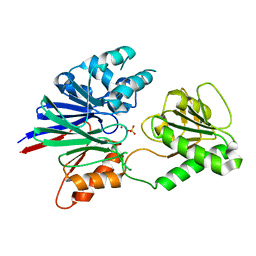 | | Structure of human CPSF-73 | | Descriptor: | Cleavage and polyadenylation specificity factor 73 kDa subunit, SULFATE ION, ZINC ION | | Authors: | Mandel, C.R, Zhang, H, Tong, L. | | Deposit date: | 2006-08-31 | | Release date: | 2007-01-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Polyadenylation factor CPSF-73 is the pre-mRNA 3'-end-processing endonuclease.
Nature, 444, 2006
|
|
2I7V
 
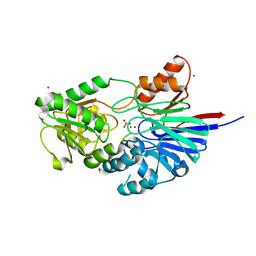 | | Structure of Human CPSF-73 | | Descriptor: | Cleavage and polyadenylation specificity factor 73 kDa subunit, SULFATE ION, ZINC ION | | Authors: | Mandel, C.R, Zhang, H, Tong, L. | | Deposit date: | 2006-08-31 | | Release date: | 2007-01-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Polyadenylation factor CPSF-73 is the pre-mRNA 3'-end-processing endonuclease.
Nature, 444, 2006
|
|
6LO2
 
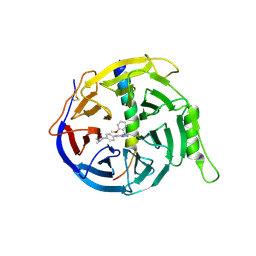 | | Crystal structure of EED in complex with EZH2 peptide and compound 11# | | Descriptor: | 8-[4-[(dimethylamino)methyl]phenyl]-N-[(2-methoxyphenyl)methyl]-[1,2,4]triazolo[4,3-a]pyridin-5-amine, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Chen, Z. | | Deposit date: | 2020-01-02 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of EED in complex with EZH2 peptide and cmpd20
To Be Published
|
|
6NIH
 
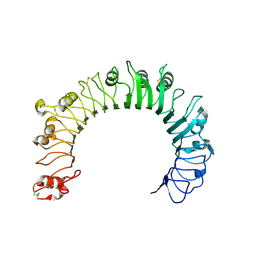 | | Crystal structure of human TLR1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Su, L, Zhang, H. | | Deposit date: | 2018-12-27 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of TLR2/TLR1 Activation by the Synthetic Agonist Diprovocim.
J. Med. Chem., 62, 2019
|
|
7WJQ
 
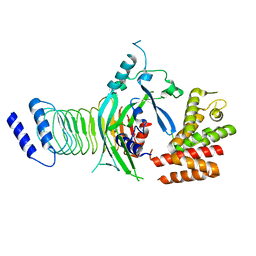 | | Crystal structure of GSDMB in complex with Ipah7.8 | | Descriptor: | Isoform 2 of Gasdermin-B, Probable E3 ubiquitin-protein ligase ipaH7.8 | | Authors: | Li, X, Zhang, H, Yin, H. | | Deposit date: | 2022-01-07 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the GSDMB-mediated cellular lysis and its targeting by IpaH7.8
Nat Commun, 14, 2023
|
|
7Y35
 
 | | Cryo-EM structure of the Abaloparatide-bound human PTH1R-Gs complex | | Descriptor: | Abaloparatide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhai, X, Mao, C, Shen, Q, Zang, S, Shen, D, Zhang, H, Chen, Z, Wang, G, Zhang, C, Zhang, Y, Liu, Z. | | Deposit date: | 2022-06-09 | | Release date: | 2023-07-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of the Abaloparatide-bound human PTH1R-Gs complex
To Be Published
|
|
6ITW
 
 | |
7CX4
 
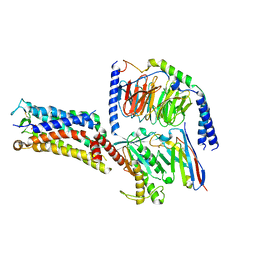 | | Cryo-EM structure of the Evatanepag-bound EP2-Gs complex | | Descriptor: | 2-[3-[[(4-~{tert}-butylphenyl)methyl-pyridin-3-ylsulfonyl-amino]methyl]phenoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
7CX3
 
 | | Cryo-EM structure of the Taprenepag-bound EP2-Gs complex | | Descriptor: | 2-[3-[[(4-pyrazol-1-ylphenyl)methyl-pyridin-3-ylsulfonyl-amino]methyl]phenoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
7CX2
 
 | | Cryo-EM structure of the PGE2-bound EP2-Gs complex | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
7WDL
 
 | | Fungal immunomodulatory protein FIP-nha | | Descriptor: | Fungal immunomodulatory protein | | Authors: | Liu, Y, Bastiaan-Net, S, Zhang, Y, Hoppenbrouwers, T, Xie, Y, Wang, Y, Wei, X, Du, G, Zhang, H, Imam, K.M.S.U, Wichers, H.J, Li, Z. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Linking the thermostability of FIP-nha (Nectria haematococca) to its structural properties.
Int.J.Biol.Macromol., 213, 2022
|
|
5VJX
 
 | | Crystal structure of the CLOCK Transcription Domain Exon19 in Complex with a Repressor | | Descriptor: | CLOCK-interacting pacemaker, Circadian locomoter output cycles protein kaput | | Authors: | Hou, Z, Su, L, Pei, J, Grishin, N.V, Zhang, H. | | Deposit date: | 2017-04-20 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Crystal Structure of the CLOCK Transactivation Domain Exon19 in Complex with a Repressor.
Structure, 25, 2017
|
|
5VJI
 
 | | Crystal structure of the CLOCK Transcription Domain Exon19 in Complex with a Repressor | | Descriptor: | CLOCK-interacting pacemaker, Circadian locomoter output cycles protein kaput | | Authors: | Hou, Z, Su, L, Pei, J, Grishin, N.V, Zhang, H. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of the CLOCK Transactivation Domain Exon19 in Complex with a Repressor.
Structure, 25, 2017
|
|
7CZ5
 
 | | Cryo-EM structure of the human growth hormone-releasing hormone receptor-Gs protein complex | | Descriptor: | CHOLESTEROL, Growth hormone-releasing hormone receptor,growth hormone-releasing hormone receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhou, F, Zhang, H, Cong, Z, Zhao, L, Zhou, Q, Mao, C, Cheng, X, Shen, D, Cai, X, Ma, C, Wang, Y, Dai, A, Zhou, Y, Sun, W, Zhao, F, Zhao, S, Jiang, H, Jiang, Y, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2020-09-07 | | Release date: | 2020-11-18 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for activation of the growth hormone-releasing hormone receptor.
Nat Commun, 11, 2020
|
|
7X7R
 
 | | Cryo-EM structure of a bacterial protein | | Descriptor: | RAMP superfamily protein, RNA (36-MER), RNA (5'-R(P*AP*GP*UP*CP*CP*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*G)-3'), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-11-16 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
