6UXN
 
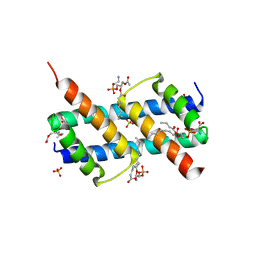 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with phosphatidylserine | | Descriptor: | Bcl-2 homologous antagonist/killer, GLYCEROL, O-[(R)-{[(2R)-2,3-bis(octanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, ... | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-11-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UXP
 
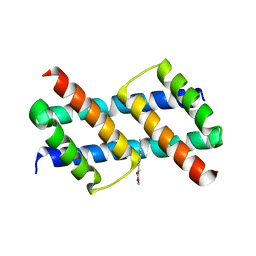 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with phosphatidylglycerol | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-[PHOSPHO-RAC-(1-GLYCEROL), Bcl-2 homologous antagonist/killer, GLYCEROL | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-11-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UXM
 
 | |
6UXR
 
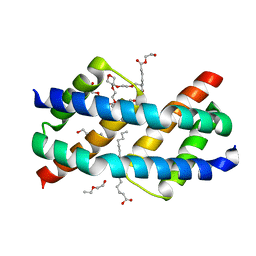 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with LysoPC | | Descriptor: | Bcl-2 homologous antagonist/killer, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-11-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UXO
 
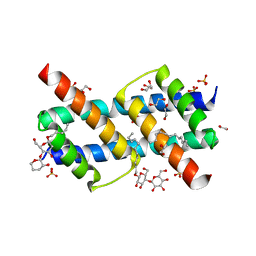 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with DDM | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Bcl-2 homologous antagonist/killer, ... | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-11-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7MX3
 
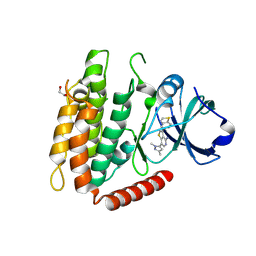 | | Crystal structure of human RIPK3 complexed with GSK'843 | | Descriptor: | 1,2-ETHANEDIOL, 3-(1,3-benzothiazol-5-yl)-7-(1,3-dimethyl-1H-pyrazol-5-yl)thieno[3,2-c]pyridin-4-amine, Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Davies, K.A, Czabotar, P.E. | | Deposit date: | 2021-05-18 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Human RIPK3 maintains MLKL in an inactive conformation prior to cell death by necroptosis.
Nat Commun, 12, 2021
|
|
5IKX
 
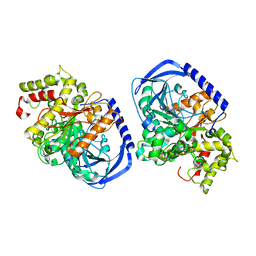 | |
6V0M
 
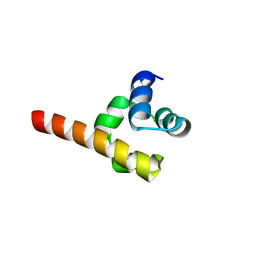 | |
6CGA
 
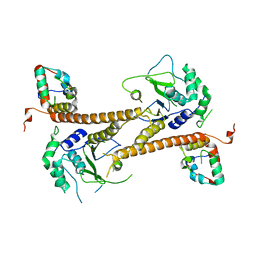 | | Structure of the PR-DUB complex | | Descriptor: | Polycomb protein Asx, Ubiquitin carboxyl-terminal hydrolase calypso | | Authors: | Foglizzo, M, Middleton, A.J, Day, C.L, Mace, P.D. | | Deposit date: | 2018-02-19 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A bidentate Polycomb Repressive-Deubiquitinase complex is required for efficient activity on nucleosomes.
Nat Commun, 9, 2018
|
|
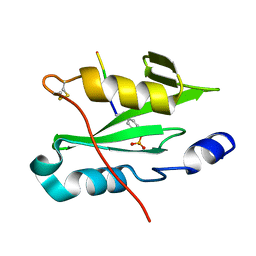
7R8W
 
 | |
7R8X
 
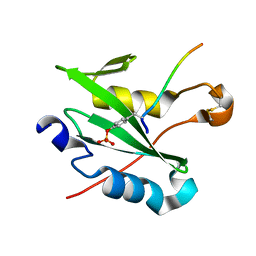 | |
6WG7
 
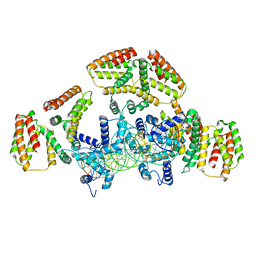 | | Coordinates of NanR dimer fitted in Hexameric NanR-DNA hetero-complex cryo-EM map | | Descriptor: | DNA (35-MER), HTH-type transcriptional repressor NanR | | Authors: | Hariprasad, V, Horne, C, Santosh, P, Amy, H, Emre, B, Rachel, N, Michael, G, Georg, R, Borries, D, Renwick, D. | | Deposit date: | 2020-04-05 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Mechanism of NanR gene repression and allosteric induction of bacterial sialic acid metabolism.
Nat Commun, 12, 2021
|
|
6WFQ
 
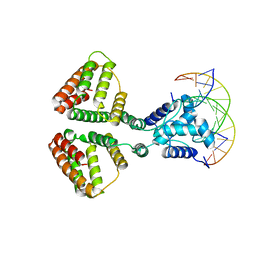 | | NanR dimer-DNA hetero-complex | | Descriptor: | DNA (5'-D(P*GP*GP*TP*AP*TP*AP*AP*CP*AP*GP*GP*TP*AP*TP*A)-3'), DNA (5'-D(P*TP*AP*TP*AP*CP*CP*TP*GP*TP*TP*AP*TP*AP*CP*C)-3'), HTH-type transcriptional repressor NanR | | Authors: | Hariprasad, V, Horne, C, Santosh, P, Amy, H, Emre, B, Rachel, N, Michael, G, Georg, R, Borries, D, Renwick, D. | | Deposit date: | 2020-04-03 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanism of NanR gene repression and allosteric induction of bacterial sialic acid metabolism.
Nat Commun, 12, 2021
|
|
4Z8N
 
 | |
6VBZ
 
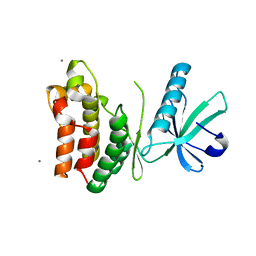 | | Crystal structure of the rat MLKL pseudokinase domain | | Descriptor: | MANGANESE (II) ION, Mixed lineage kinase domain-like pseudokinase | | Authors: | Davies, K.A, Czabotar, P.E. | | Deposit date: | 2019-12-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Distinct pseudokinase domain conformations underlie divergent activation mechanisms among vertebrate MLKL orthologues.
Nat Commun, 11, 2020
|
|
6VC0
 
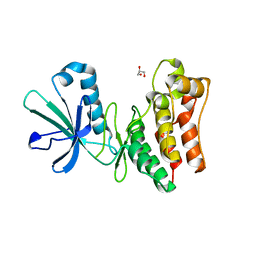 | | Crystal structure of the horse MLKL pseudokinase domain | | Descriptor: | GLYCEROL, Mixed lineage kinase domain like pseudokinase | | Authors: | Davies, K.A, Czabotar, P.E. | | Deposit date: | 2019-12-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.746 Å) | | Cite: | Distinct pseudokinase domain conformations underlie divergent activation mechanisms among vertebrate MLKL orthologues.
Nat Commun, 11, 2020
|
|
6BPD
 
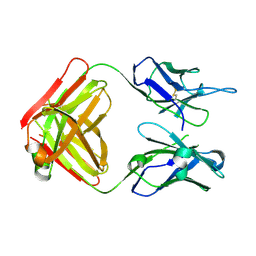 | |
6D03
 
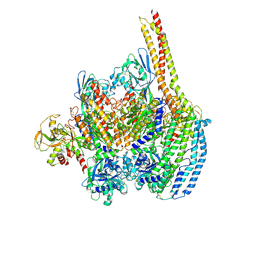 | | Cryo-EM structure of a Plasmodium vivax invasion complex essential for entry into human reticulocytes; one molecule of parasite ligand. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(2-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruszczyk, J, Huang, R.K, Hong, C, Yu, Z, Tham, W.H. | | Deposit date: | 2018-04-10 | | Release date: | 2018-06-20 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
5VE6
 
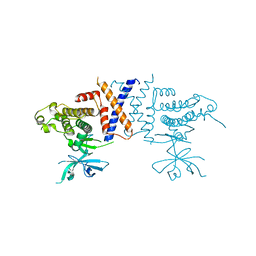 | | Crystal structure of Sugen kinase 223 | | Descriptor: | Tyrosine-protein kinase SgK223 | | Authors: | Patel, O, Lucet, I, Panjikar, S. | | Deposit date: | 2017-04-03 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.953 Å) | | Cite: | Structure of SgK223 pseudokinase reveals novel mechanisms of homotypic and heterotypic association.
Nat Commun, 8, 2017
|
|
6D05
 
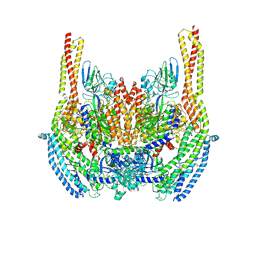 | | Cryo-EM structure of a Plasmodium vivax invasion complex essential for entry into human reticulocytes; two molecules of parasite ligand, subclass 2. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Gruszczyk, J, Huang, R.K, Hong, C, Yu, Z, Tham, W.H. | | Deposit date: | 2018-04-10 | | Release date: | 2018-06-20 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
6D04
 
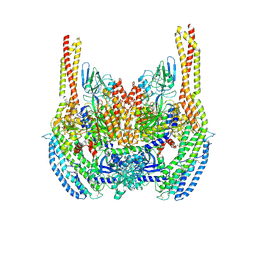 | | Cryo-EM structure of a Plasmodium vivax invasion complex essential for entry into human reticulocytes; two molecules of parasite ligand, subclass 1. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Gruszczyk, J, Huang, R.K, Hong, C, Yu, Z, Tham, W.H. | | Deposit date: | 2018-04-10 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
6C5X
 
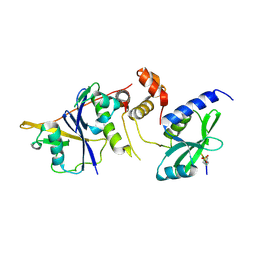 | | Crystal Structure of SOCS1 in complex with ElonginB and ElonginC | | Descriptor: | Elongin-B, Elongin-C, GP130 peptide fragment, ... | | Authors: | Kershaw, N.J, Laktyushin, A, Babon, J.J. | | Deposit date: | 2018-01-17 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.105 Å) | | Cite: | The molecular basis of JAK/STAT inhibition by SOCS1.
Nat Commun, 9, 2018
|
|
6BPA
 
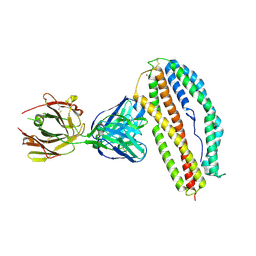 | | Plasmodium vivax reticulocyte binding protein 2b (PvRBP2b) bound to monoclonal antibody 3E9 | | Descriptor: | BROMIDE ION, Monoclonal antibody 3E9 Fab heavy chain, Monoclonal antibody 3E9 Fab light chain, ... | | Authors: | Gruszczyk, J, Chan, L.J, Tham, W.H. | | Deposit date: | 2017-11-22 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
6BPC
 
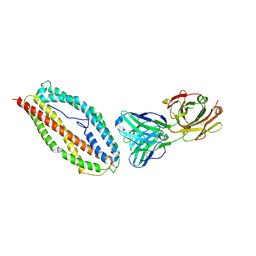 | | Plasmodium vivax reticulocyte binding protein 2b (PvRBP2b) bound to monoclonal antibody 4F7 | | Descriptor: | Monoclonal antibody 4F7 Fab heavy chain, Monoclonal antibody 4F7 Fab light chain, Reticulocyte binding protein 2, ... | | Authors: | Gruszczyk, J, Chan, L.J, Tham, W.H. | | Deposit date: | 2017-11-22 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
6BPB
 
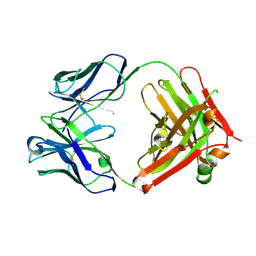 | |
