5VNP
 
 | |
7XTQ
 
 | | Cryo-EM structure of the R399-bound GPBAR-Gs complex | | Descriptor: | G-protein coupled bile acid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ma, L, Yang, F, Wu, X, Mao, C, Sun, J, Yu, X, Zhang, Y, Zhang, P. | | Deposit date: | 2022-05-17 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis and molecular mechanism of biased GPBAR signaling in regulating NSCLC cell growth via YAP activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4PY4
 
 | |
4QPL
 
 | | Crystal structure of RNF146(RING-WWE)/UbcH5a/iso-ADPr complex | | Descriptor: | 2'-O-(5-O-phosphono-alpha-D-ribofuranosyl)adenosine 5'-(dihydrogen phosphate), E3 ubiquitin-protein ligase RNF146, Ubiquitin-conjugating enzyme E2 D1, ... | | Authors: | Wang, Z, DaRosa, P.A, Klevit, R.E, Xu, W. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal.
Nature, 517, 2015
|
|
7Y9G
 
 | | Crystal structure of diterpene synthase VenA from Streptomyces venezuelae ATCC 15439 in complex with pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, L.L, Xie, Z.Z, Huang, J.-W, Hu, Y.C, Liu, W.D, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase.
Nat Commun, 14, 2023
|
|
7Y9H
 
 | | Crystal structure of diterpene synthase VenA from Streptomyces venezuelae ATCC 15439 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Diterpene synthase VenA, ... | | Authors: | Zhang, L.L, Xie, Z.Z, Huang, J.-W, Hu, Y.C, Liu, W.D, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase.
Nat Commun, 14, 2023
|
|
8IL3
 
 | | Cryo-EM structure of CD38 in complex with FTL004 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, Heavy chain, Light chain | | Authors: | Yang, J, Wang, Y, Zhang, G. | | Deposit date: | 2023-03-01 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | FTL004, an anti-CD38 mAb with negligible RBC binding and enhanced pro-apoptotic activity, is a novel candidate for treatments of multiple myeloma and non-Hodgkin lymphoma.
J Hematol Oncol, 15, 2022
|
|
5ZNK
 
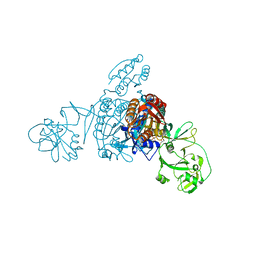 | | Crystal structure of a bacterial ProRS with ligands | | Descriptor: | 7-chloro-6-fluoro-3-{2-oxo-3-[(2S)-piperidin-2-yl]propyl}quinazolin-4(3H)-one, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cheng, B, Yu, Y, Zhou, H. | | Deposit date: | 2018-04-09 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Design of Halofuginone Derivatives as ATP-Aided Inhibitors Against Bacterial Prolyl-tRNA Synthetase.
J.Med.Chem., 65, 2022
|
|
6BCU
 
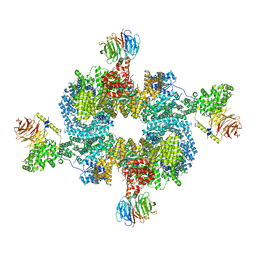 | |
6BCX
 
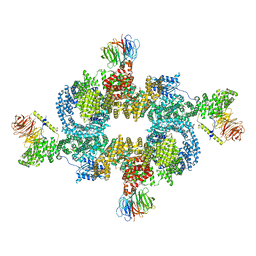 | | mTORC1 structure refined to 3.0 angstroms | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E-binding protein 1, MAGNESIUM ION, ... | | Authors: | Pavletich, N.P, Yang, H. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40.
Nature, 552, 2017
|
|
6AVO
 
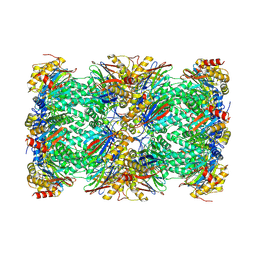 | | Cryo-EM structure of human immunoproteasome with a novel noncompetitive inhibitor that selectively inhibits activated lymphocytes | | Descriptor: | N~1~-{2-[([1,1'-biphenyl]-3-carbonyl)amino]ethyl}-N~4~-tert-butyl-N~2~-(3-phenylpropanoyl)-L-aspartamide, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Li, H, Santos, R, Bai, L. | | Deposit date: | 2017-09-04 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of human immunoproteasome with a reversible and noncompetitive inhibitor that selectively inhibits activated lymphocytes.
Nat Commun, 8, 2017
|
|
6CMX
 
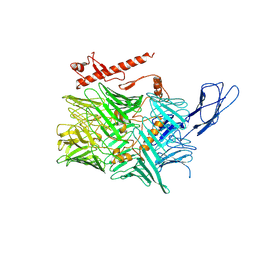 | | Human Teneurin 2 extra-cellular region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Teneurin-2, alpha-D-mannopyranose, ... | | Authors: | Shalev-Benami, M, Li, J, Sudhof, T, Skiniotis, G, Arac, D. | | Deposit date: | 2018-03-06 | | Release date: | 2018-07-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis for Teneurin Function in Circuit-Wiring: A Toxin Motif at the Synapse.
Cell, 173, 2018
|
|
5ZNJ
 
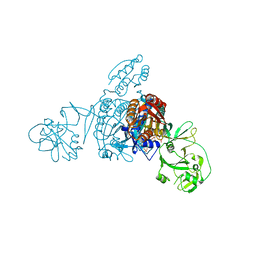 | | Crystal structure of a bacterial ProRS with ligands | | Descriptor: | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cheng, B, Yu, Y, Zhou, H. | | Deposit date: | 2018-04-09 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-Guided Design of Halofuginone Derivatives as ATP-Aided Inhibitors Against Bacterial Prolyl-tRNA Synthetase.
J.Med.Chem., 65, 2022
|
|
6CYW
 
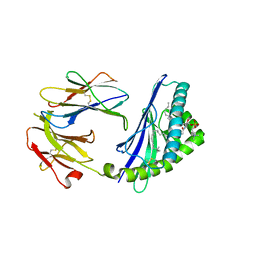 | | Structure of sphingomyelin in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-04-06 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Control of CD1d-restricted antigen presentation and inflammation by sphingomyelin.
Nat.Immunol., 20, 2019
|
|
6M2B
 
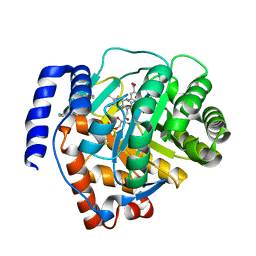 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with S416 | | Descriptor: | 2-[(E)-[[4-(2-chlorophenyl)-1,3-thiazol-2-yl]-methyl-hydrazinylidene]methyl]benzoic acid, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Li, H. | | Deposit date: | 2020-02-27 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Novel and potent inhibitors targeting DHODH are broad-spectrum antivirals against RNA viruses including newly-emerged coronavirus SARS-CoV-2.
Protein Cell, 11, 2020
|
|
6VBG
 
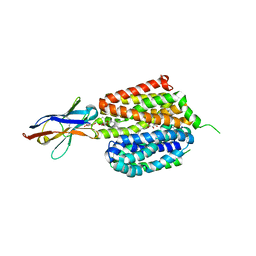 | | Lactose permease complex with thiodigalactoside and nanobody 9043 | | Descriptor: | Galactoside permease, beta-D-galactopyranose-(1-1)-1-thio-beta-D-galactopyranose, nanobody 9043, ... | | Authors: | Kumar, H, Stroud, R.M, Kaback, H.R, Finer-Moore, J, Smirnova, I, Kasho, V, Pardon, E, Steyart, J. | | Deposit date: | 2019-12-18 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Diversity in kinetics correlated with structure in nano body-stabilized LacY.
Plos One, 15, 2020
|
|
7KMY
 
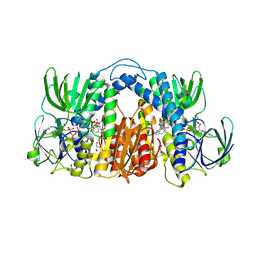 | | Structure of Mtb Lpd bound to 010705 | | Descriptor: | Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Lima, C.D. | | Deposit date: | 2020-11-03 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Whole Cell Active Inhibitors of Mycobacterial Lipoamide Dehydrogenase Afford Selectivity over the Human Enzyme through Tight Binding Interactions.
Acs Infect Dis., 7, 2021
|
|
7KI5
 
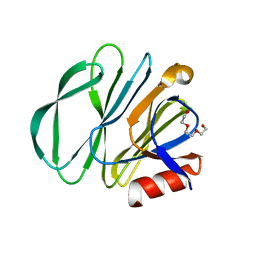 | | Crystal structure of P[6] rotavirus vp8* in complex with LNT | | Descriptor: | Capsid protein, GLYCEROL, TRIETHYLENE GLYCOL, ... | | Authors: | Xu, S, Kennedy, M.A. | | Deposit date: | 2020-10-23 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural basis of P[II] rotavirus evolution and host ranges under selection of histo-blood group antigens.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DPV
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with 7-O-methyl-dihydromyricetin | | Descriptor: | (2S,3S)-3,5-dihydroxy-7-methoxy-2-(3,4,5-trihydroxyphenyl)chroman-4-one, 3C-like proteinase | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
7DPU
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with 7-O-methyl-myricetin | | Descriptor: | 3C-like proteinase, 7-methoxy-3,5-bis(oxidanyl)-2-[3,4,5-tris(oxidanyl)phenyl]chromen-4-one, GLYCEROL | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
7DPP
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with myricetin | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, 3C-like proteinase | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
8YM1
 
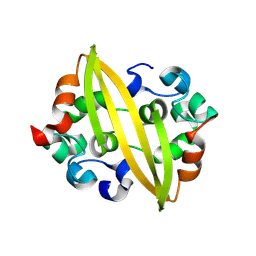 | | Structure of SADS-CoV Virus Nucleocapsid Protein | | Descriptor: | nucleocapsid phosphoprotein | | Authors: | Zhang, Y, Wu, F, Xu, W. | | Deposit date: | 2024-03-08 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unraveling the assembly mechanism of SADS-CoV virus nucleocapsid protein: insights from RNA binding, dimerization, and epitope diversity profiling.
J.Virol., 2024
|
|
8JIY
 
 | | A carbohydrate binding domain of a putative chondroitinase | | Descriptor: | DUF4955 domain-containing protein | | Authors: | Liu, G.C, Chang, Y.G. | | Deposit date: | 2023-05-29 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and structural characterization of a novel chondroitin sulfate-specific carbohydrate-binding module: The first member of a new family, CBM100.
Int.J.Biol.Macromol., 255, 2024
|
|
5WBH
 
 | |
5WBY
 
 | |
