5T2A
 
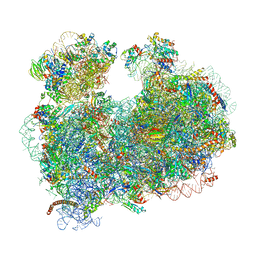 | | CryoEM structure of the Leishmania donovani 80S ribosome at 2.9 Angstrom resolution | | 分子名称: | 18S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Zhang, X, Lai, M, Zhou, Z.H. | | 登録日 | 2016-08-23 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes.
Nat Commun, 7, 2016
|
|
5T2C
 
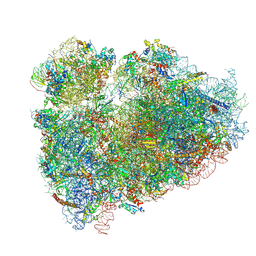 | | CryoEM structure of the human ribosome at 3.6 Angstrom resolution | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | 著者 | Zhang, X, Lai, M, Zhou, Z.H. | | 登録日 | 2016-08-23 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes.
Nat Commun, 7, 2016
|
|
8UEL
 
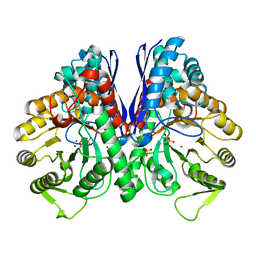 | |
1ML9
 
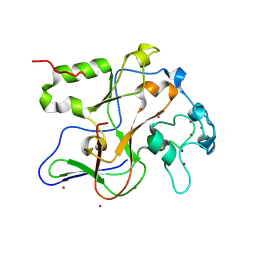 | | Structure of the Neurospora SET domain protein DIM-5, a histone lysine methyltransferase | | 分子名称: | Histone H3 methyltransferase DIM-5, UNKNOWN LIGAND, ZINC ION | | 著者 | Zhang, X, Tamaru, H, Khan, S.I, Horton, J.R, Keefe, L.J, Selker, E.U, Cheng, X. | | 登録日 | 2002-08-30 | | 公開日 | 2002-10-23 | | 最終更新日 | 2025-02-12 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structure of the Neurospora SET domain protein DIM-5,
a histone H3 lysine methyltransferase
Cell(Cambridge,Mass.), 111, 2002
|
|
5XJC
 
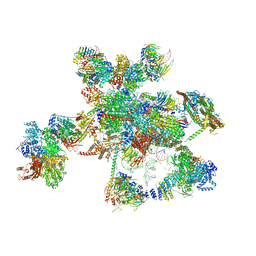 | | Cryo-EM structure of the human spliceosome just prior to exon ligation at 3.6 angstrom | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Zhang, X, Yan, C, Hang, J, Finci, I.L, Lei, J, Shi, Y. | | 登録日 | 2017-04-30 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | An Atomic Structure of the Human Spliceosome
Cell, 169, 2017
|
|
8HMY
 
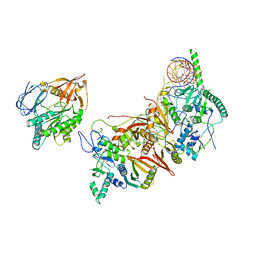 | | Cryo-EM structure of the human pre-catalytic TSEN/pre-tRNA complex | | 分子名称: | Chromosome 1 open reading frame 19, isoform CRA_a, MAGNESIUM ION, ... | | 著者 | Zhang, X, Yang, F, Zhan, X, Shi, Y. | | 登録日 | 2022-12-06 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | Structural basis of pre-tRNA intron removal by human tRNA splicing endonuclease.
Mol.Cell, 83, 2023
|
|
7VPX
 
 | |
8HMZ
 
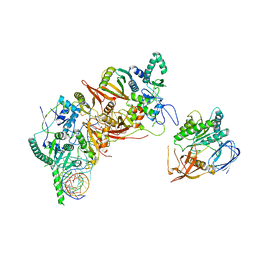 | | Cryo-EM structure of the human post-catalytic TSEN/pre-tRNA complex | | 分子名称: | Chromosome 1 open reading frame 19, isoform CRA_a, MAGNESIUM ION, ... | | 著者 | Zhang, X, Yang, F, Zhan, X, Shi, Y. | | 登録日 | 2022-12-06 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis of pre-tRNA intron removal by human tRNA splicing endonuclease.
Mol.Cell, 83, 2023
|
|
1PEG
 
 | | Structural basis for the product specificity of histone lysine methyltransferases | | 分子名称: | Histone H3, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | 著者 | Zhang, X, Yang, Z, Khan, S.I, Horton, J.R, Tamaru, H, Selker, E.U, Cheng, X. | | 登録日 | 2003-05-21 | | 公開日 | 2003-08-05 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Structural basis for the product specificity of histone lysine methyltransferases
Mol.Cell, 12, 2003
|
|
7W5A
 
 | | The cryo-EM structure of human pre-C*-II complex | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | 著者 | Zhan, X, Lu, Y, Shi, Y. | | 登録日 | 2021-11-29 | | 公開日 | 2022-06-22 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Mechanism of exon ligation by human spliceosome.
Mol.Cell, 82, 2022
|
|
7W5B
 
 | | The cryo-EM structure of human C* complex | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | 著者 | Zhan, X, Lu, Y, Shi, Y. | | 登録日 | 2021-11-29 | | 公開日 | 2022-06-22 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Mechanism of exon ligation by human spliceosome.
Mol.Cell, 82, 2022
|
|
7W59
 
 | | The cryo-EM structure of human pre-C*-I complex | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | 著者 | Zhan, X, Lu, Y, Shi, Y. | | 登録日 | 2021-11-29 | | 公開日 | 2022-06-22 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Mechanism of exon ligation by human spliceosome.
Mol.Cell, 82, 2022
|
|
5TEC
 
 | | Crystal structure of the TIR domain from the Arabidopsis thaliana NLR protein SNC1 | | 分子名称: | Protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1 | | 著者 | Zhang, X, Bentham, A, Ve, T, Williams, S.J, Kobe, B. | | 登録日 | 2016-09-20 | | 公開日 | 2017-02-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Multiple functional self-association interfaces in plant TIR domains.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2Y3A
 
 | | Crystal structure of p110beta in complex with icSH2 of p85beta and the drug GDC-0941 | | 分子名称: | 2-(1H-indazol-4-yl)-6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-yl-thieno[3,2-d]pyrimidine, PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY SUBUNIT BETA, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT BETA ISOFORM | | 著者 | Zhang, X, Vadas, O, Perisic, O, Williams, R.L. | | 登録日 | 2010-12-20 | | 公開日 | 2011-03-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structure of Lipid Kinase P110Beta-P85Beta Elucidates an Unusual Sh2-Domain-Mediated Inhibitory Mechanism.
Mol.Cell, 41, 2011
|
|
5GTX
 
 | | Crystal structure of mutated buckwheat glutaredoxin | | 分子名称: | buckwheat glutaredoxin | | 著者 | Zhang, X, Wang, W, Zhao, Y, Wang, Z, Wang, H. | | 登録日 | 2016-08-23 | | 公開日 | 2017-07-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Structural insights into the binding of buckwheat glutaredoxin with GSH and regulation of its catalytic activity
J. Inorg. Biochem., 173, 2017
|
|
5KQA
 
 | | Crystal structure of buckwheat glutaredoxin-glutathione complex | | 分子名称: | GLUTATHIONE, Glutaredoxin-glutathione complex | | 著者 | Zhang, X, Wang, W, Zhao, Y, Wang, Z, Wang, H. | | 登録日 | 2016-07-06 | | 公開日 | 2017-07-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural insights into the binding of buckwheat glutaredoxin with GSH and regulation of its catalytic activity
J. Inorg. Biochem., 173, 2017
|
|
9IMU
 
 | |
8T3V
 
 | | Cryo-EM structure of the DHA bound FFA1-Gq complex | | 分子名称: | CHOLESTEROL, DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 1, ... | | 著者 | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | 登録日 | 2023-06-07 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
8T3O
 
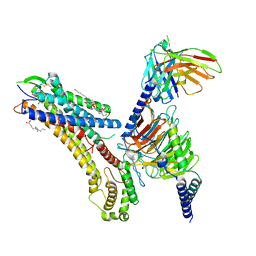 | | Cryo-EM structure of the TUG-891 bound FFA4-Gq complex | | 分子名称: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, ... | | 著者 | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | 登録日 | 2023-06-07 | | 公開日 | 2024-01-17 | | 最終更新日 | 2025-05-14 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
8T3Q
 
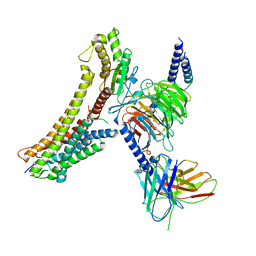 | | Cryo-EM structure of the DHA bound FFA4-Gq complex | | 分子名称: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | 登録日 | 2023-06-07 | | 公開日 | 2024-01-24 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
8T3S
 
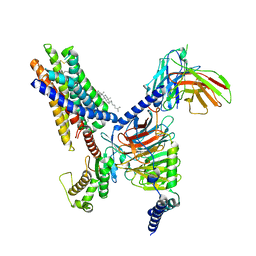 | | Cryo-EM structure of the Butyrate bound FFA2-Gq complex | | 分子名称: | CHOLESTEROL, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | 登録日 | 2023-06-07 | | 公開日 | 2024-01-24 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
5Z57
 
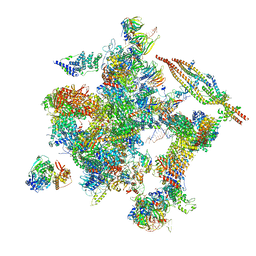 | | Cryo-EM structure of the human activated spliceosome (late Bact) at 6.5 angstrom | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ALANINE, BUD13 homolog, ... | | 著者 | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | 登録日 | 2018-01-17 | | 公開日 | 2018-09-19 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
5YZG
 
 | | The Cryo-EM Structure of Human Catalytic Step I Spliceosome (C complex) at 4.1 angstrom resolution | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Zhan, X, Yan, C, Zhang, X, Lei, J, Shi, Y. | | 登録日 | 2017-12-14 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structure of a human catalytic step I spliceosome
Science, 359, 2018
|
|
3F4M
 
 | | Crystal structure of TIPE2 | | 分子名称: | CHLORIDE ION, Tumor necrosis factor, alpha-induced protein 8-like protein 2 | | 著者 | Zhang, X, Wang, J, Shi, Y. | | 登録日 | 2008-11-01 | | 公開日 | 2009-03-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.696 Å) | | 主引用文献 | Crystal structure of TIPE2 provides insights into immune homeostasis
Nat.Struct.Mol.Biol., 16, 2009
|
|
5Z56
 
 | | cryo-EM structure of a human activated spliceosome (mature Bact) at 5.1 angstrom. | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | 著者 | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | 登録日 | 2018-01-17 | | 公開日 | 2018-09-19 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
