6QEO
 
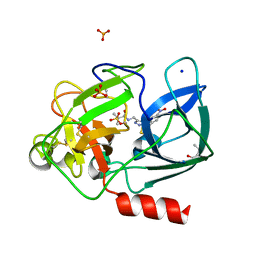 | | Crystal structure of Porcine Pancreatic Elastase (PPE) in complex with the 3-Oxo-beta-Sultam inhibitor LMC269 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-methyl-1-[[1-[(4-nitrophenyl)methyl]-1,2,3-triazol-4-yl]methylamino]-1-oxidanylidene-propane-2-sulfonic acid, Chymotrypsin-like elastase family member 1, ... | | 著者 | Brito, J.A, Almeida, V.T, Carvalho, L.M, Moreira, R, Archer, M. | | 登録日 | 2019-01-08 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | 3-Oxo-beta-sultam as a Sulfonylating Chemotype for Inhibition of Serine Hydrolases and Activity-Based Protein Profiling.
Acs Chem.Biol., 15, 2020
|
|
6SMA
 
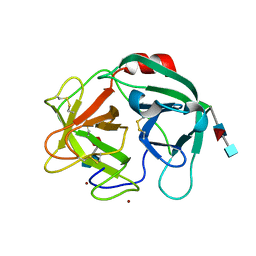 | | Crystal structure of Human Neutrophil Elastase (HNE) in complex with the 3-Oxo-beta-Sultam inhibitor LMC249 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[[1-[(4-bromophenyl)methyl]-1,2,3-triazol-4-yl]methylcarbamoyl]pentane-3-sulfonic acid, ... | | 著者 | Brito, J.A, Almeida, V.T, Carvalho, L.M, Moreira, R, Archer, M. | | 登録日 | 2019-08-21 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | 3-Oxo-beta-sultam as a Sulfonylating Chemotype for Inhibition of Serine Hydrolases and Activity-Based Protein Profiling.
Acs Chem.Biol., 15, 2020
|
|
6QBU
 
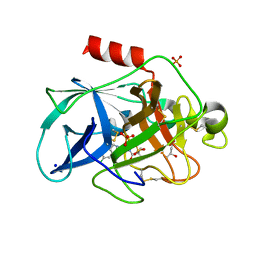 | | Crystal structure of Porcine Pancreatic Elastase (PPE) in complex with the 3-Oxo-beta-Sultam inhibitor LMC188 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Chymotrypsin-like elastase family member 1, PHOSPHATE ION, ... | | 著者 | Brito, J.A, Almeida, V.T, Carvalho, L.M, Moreira, R, Archer, M. | | 登録日 | 2018-12-21 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | 3-Oxo-beta-sultam as a Sulfonylating Chemotype for Inhibition of Serine Hydrolases and Activity-Based Protein Profiling.
Acs Chem.Biol., 15, 2020
|
|
7A3F
 
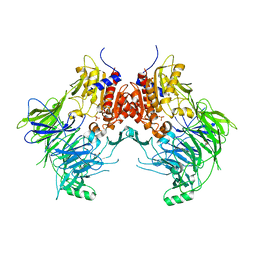 | | Crystal structure of apo DPP9 | | 分子名称: | Dipeptidyl peptidase 9, GLYCEROL, PHOSPHATE ION | | 著者 | Ross, B.H, Huber, R. | | 登録日 | 2020-08-18 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Discovery and Development of 4-Oxo-beta-Lactams as Novel Inhibitors of Dipeptidyl Peptidases 8 and 9
To Be Published
|
|
7A3G
 
 | | Crystal structure of DPP8 in complex with a 4-oxo-b-lactam based inhibitor, 91 | | 分子名称: | 1-[3-(7,8-dihydro-5~{H}-[1,3]dioxolo[4,5-g]isoquinolin-6-ylmethyl)phenyl]-3,3-diethyl-azetidine-2,4-dione, CHLORIDE ION, Dipeptidyl peptidase 8, ... | | 著者 | Ross, B.H, Huber, R. | | 登録日 | 2020-08-18 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Chemoproteomics-Enabled Identification of 4-Oxo-beta-Lactams as Inhibitors of Dipeptidyl Peptidases 8 and 9.
Angew.Chem.Int.Ed.Engl., 2022
|
|
7A3L
 
 | |
7A3J
 
 | |
7A3I
 
 | |
7A3K
 
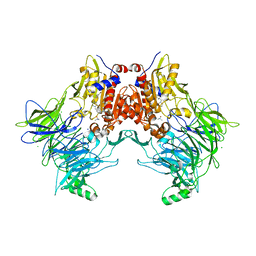 | |
7AYR
 
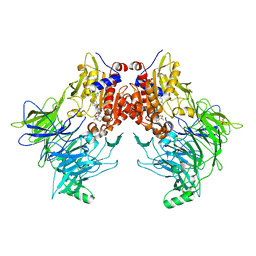 | |
7AYQ
 
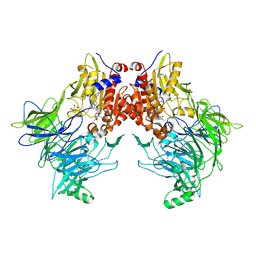 | |
2WAP
 
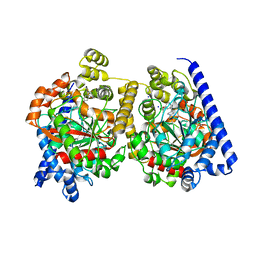 | | 3D-crystal structure of humanized-rat fatty acid amide hydrolase (FAAH) conjugated with the drug-like urea inhibitor PF-3845 | | 分子名称: | 4-(3-{[5-(trifluoromethyl)pyridin-2-yl]oxy}benzyl)piperidine-1-carboxylic acid, CHLORIDE ION, FATTY-ACID AMIDE HYDROLASE 1, ... | | 著者 | Mileni, M, Kamtekar, S, Stevens, R.C. | | 登録日 | 2009-02-11 | | 公開日 | 2009-05-05 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery and Characterization of a Highly Selective Faah Inhibitor that Reduces Inflammatory Pain.
Chem.Biol., 16, 2009
|
|
4JLL
 
 | | Crystal Structure of the evolved variant of the computationally designed serine hydrolase, OSH55.4_H1 covalently bound with FP-alkyne, Northeast Structural Genomics Consortium (NESG) Target OR273 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Tong, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-03-12 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4JVV
 
 | | Crystal structure of the evolved variant of the computationally designed serine hydrolase, OSH55.4_H1, covalently bound with diisopropyl fluorophosphate (DFP), Northeast Structural Genomics Consortium (NESG) Target OR273 | | 分子名称: | evolved variant of the computationally designed serine hydrolase | | 著者 | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Tong, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-03-26 | | 公開日 | 2013-04-24 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.288 Å) | | 主引用文献 | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4JCA
 
 | | Crystal Structure of the apo form of the evolved variant of the computationally designed serine hydrolase, OSH55.4_H1. Northeast Structural Genomics Consortium (NESG) Target OR273 | | 分子名称: | CITRIC ACID, RUBIDIUM ION, serine hydrolase | | 著者 | Kuzin, A.P, Lew, S, Rajagopalan, S, Seetharaman, J, Tong, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-02-21 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.411 Å) | | 主引用文献 | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
3LJ6
 
 | |
3LJ7
 
 | |
3K84
 
 | | Crystal Structure Analysis of a Oleyl/Oxadiazole/pyridine Inhibitor Bound to a Humanized Variant of Fatty Acid Amide Hydrolase | | 分子名称: | (9Z)-1-(5-pyridin-2-yl-1,3,4-oxadiazol-2-yl)octadec-9-en-1-one, CHLORIDE ION, Fatty-acid amide hydrolase 1 | | 著者 | Mileni, M, Stevens, R.C, Boger, D.L. | | 登録日 | 2009-10-13 | | 公開日 | 2009-12-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | X-ray crystallographic analysis of alpha-ketoheterocycle inhibitors bound to a humanized variant of fatty acid amide hydrolase.
J.Med.Chem., 53, 2010
|
|
3K83
 
 | | Crystal Structure Analysis of a Biphenyl/Oxazole/Carboxypyridine alpha-ketoheterocycle Inhibitor Bound to a Humanized Variant of Fatty Acid Amide Hydrolase | | 分子名称: | 1-DODECANOL, 6-[2-(3-biphenyl-4-ylpropanoyl)-1,3-oxazol-5-yl]pyridine-2-carboxylic acid, CHLORIDE ION, ... | | 著者 | Mileni, M, Stevens, R.C, Boger, D.L. | | 登録日 | 2009-10-13 | | 公開日 | 2009-12-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.251 Å) | | 主引用文献 | X-ray crystallographic analysis of alpha-ketoheterocycle inhibitors bound to a humanized variant of fatty acid amide hydrolase.
J.Med.Chem., 53, 2010
|
|
3K7F
 
 | | Crystal Structure Analysis of a Phenhexyl/Oxazole/Carboxypyridine alpha-Ketoheterocycle Inhibitor Bound to a Humanized Variant of Fatty Acid Amide Hydrolase' | | 分子名称: | 6-[2-(7-phenylheptanoyl)-1,3-oxazol-5-yl]pyridine-2-carboxylic acid, CHLORIDE ION, Fatty-acid amide hydrolase 1, ... | | 著者 | Mileni, M, Stevens, R.C, Boger, D.L. | | 登録日 | 2009-10-13 | | 公開日 | 2009-12-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | X-ray crystallographic analysis of alpha-ketoheterocycle inhibitors bound to a humanized variant of fatty acid amide hydrolase.
J.Med.Chem., 53, 2010
|
|
5UI4
 
 | | Structure of NME1 covalently conjugated to imidazole fluorosulfate | | 分子名称: | 4-[4-(3-methoxyphenyl)-1-(prop-2-yn-1-yl)-1H-imidazol-5-yl]phenyl sulfurofluoridate, Nucleoside diphosphate kinase A | | 著者 | Mortenson, D.E, Brighty, G.J, Wilson, I.A, Kelly, J.W. | | 登録日 | 2017-01-12 | | 公開日 | 2018-01-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | "Inverse Drug Discovery" Strategy To Identify Proteins That Are Targeted by Latent Electrophiles As Exemplified by Aryl Fluorosulfates.
J. Am. Chem. Soc., 140, 2018
|
|
5UEH
 
 | | Structure of GSTO1 covalently conjugated to quinolinic acid fluorosulfate | | 分子名称: | 2-(4-chlorophenyl)-6-[(fluorosulfonyl)oxy]quinoline-4-carboxylic acid, GLYCEROL, Glutathione S-transferase omega-1, ... | | 著者 | Mortenson, D.E, Wilson, I.A, Kelly, J.W. | | 登録日 | 2017-01-02 | | 公開日 | 2018-01-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | "Inverse Drug Discovery" Strategy To Identify Proteins That Are Targeted by Latent Electrophiles As Exemplified by Aryl Fluorosulfates.
J. Am. Chem. Soc., 140, 2018
|
|
5UWD
 
 | |
