7Q51
 
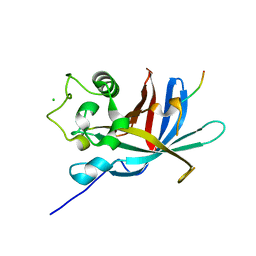 | | yeast Gid10 bound to a Phe/N-peptide | | 分子名称: | CHLORIDE ION, FWLPANLW peptide, Uncharacterized protein YGR066C | | 著者 | Chrustowicz, J, Sherpa, D, Prabu, J.R, Schulman, B.A. | | 登録日 | 2021-11-02 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Multifaceted N-Degron Recognition and Ubiquitylation by GID/CTLH E3 Ligases.
J.Mol.Biol., 434, 2022
|
|
7Q50
 
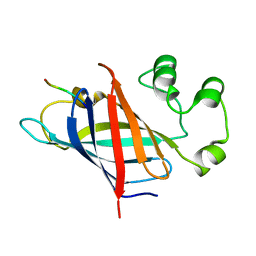 | | human Gid4 bound to a Phe/N-peptide | | 分子名称: | FDVSWFMG peptide, Glucose-induced degradation protein 4 homolog | | 著者 | Chrustowicz, J, Sherpa, D, Loke, M.S, Prabu, J.R, Schulman, B.A. | | 登録日 | 2021-11-02 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.16 Å) | | 主引用文献 | Multifaceted N-Degron Recognition and Ubiquitylation by GID/CTLH E3 Ligases.
J.Mol.Biol., 434, 2022
|
|
8CDK
 
 | |
8CDJ
 
 | |
8CAF
 
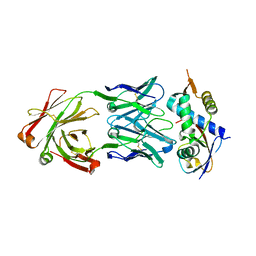 | | N8C_Fab3b in complex with NEDD8-CUL1(WHB) | | 分子名称: | Cullin-1, Fab Heavy Chain, Fab Light Chain, ... | | 著者 | Duda, D.M, Yanishevski, D, Henneberg, L.T, Schulman, B.A. | | 登録日 | 2023-01-24 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Activity-based profiling of cullin-RING E3 networks by conformation-specific probes.
Nat.Chem.Biol., 19, 2023
|
|
8C07
 
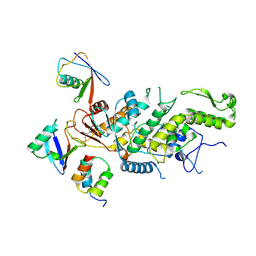 | |
8C06
 
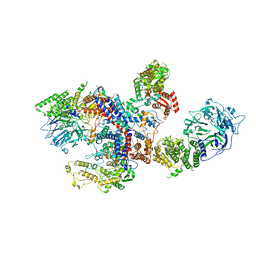 | |
4YII
 
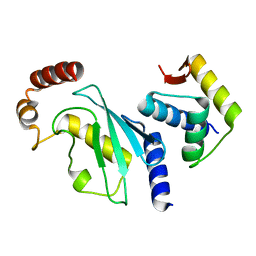 | |
7ONI
 
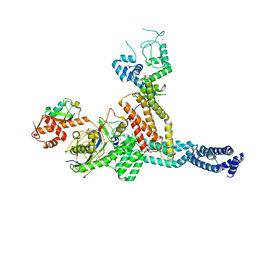 | | Structure of Neddylated CUL5 C-terminal region-RBX2-ARIH2* | | 分子名称: | Cullin-5, E3 ubiquitin-protein ligase ARIH2, NEDD8, ... | | 著者 | Kostrhon, S.P, prabu, J.R, Schulman, B.A. | | 登録日 | 2021-05-25 | | 公開日 | 2021-09-15 | | 最終更新日 | 2021-10-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | CUL5-ARIH2 E3-E3 ubiquitin ligase structure reveals cullin-specific NEDD8 activation.
Nat.Chem.Biol., 17, 2021
|
|
8OIF
 
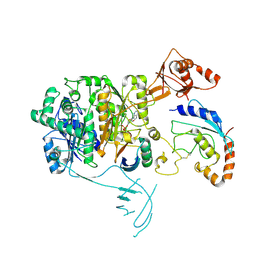 | | Structure of the UBE1L activating enzyme bound to ISG15 and UBE2L6 | | 分子名称: | ADENOSINE MONOPHOSPHATE, Ubiquitin-like modifier-activating enzyme 7, Ubiquitin-like protein ISG15, ... | | 著者 | Wallace, I, Kheewoong, B, Prabu, J.R, Vollrath, R, von Gronau, S, Schulman, B.A, Swatek, K.N. | | 登録日 | 2023-03-22 | | 公開日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Insights into the ISG15 transfer cascade by the UBE1L activating enzyme.
Nat Commun, 14, 2023
|
|
8PMQ
 
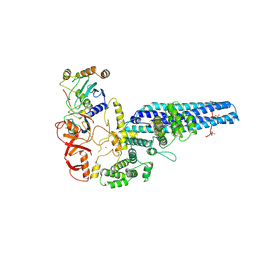 | | Catalytic module of yeast GID E3 ligase bound to multiphosphorylated Ubc8~ubiquitin | | 分子名称: | E3 ubiquitin-protein ligase RMD5, Protein FYV10, Ubiquitin, ... | | 著者 | Chrustowicz, J, Sherpa, D, Prabu, R.J, Schulman, B.A. | | 登録日 | 2023-06-29 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.53 Å) | | 主引用文献 | Multisite phosphorylation dictates selective E2-E3 pairing as revealed by Ubc8/UBE2H-GID/CTLH assemblies.
Mol.Cell, 84, 2024
|
|
8PJN
 
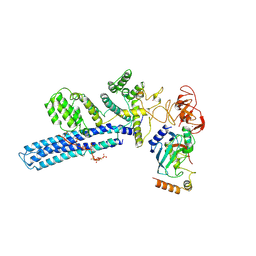 | | Catalytic module of human CTLH E3 ligase bound to multiphosphorylated UBE2H~ubiquitin | | 分子名称: | E3 ubiquitin-protein transferase MAEA, E3 ubiquitin-protein transferase RMND5A, Ubiquitin, ... | | 著者 | Chrustowicz, J, Sherpa, D, Prabu, R.J, Schulman, B.A. | | 登録日 | 2023-06-23 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Multisite phosphorylation dictates selective E2-E3 pairing as revealed by Ubc8/UBE2H-GID/CTLH assemblies.
Mol.Cell, 84, 2024
|
|
8PDA
 
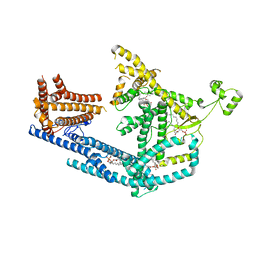 | | cryo-EM structure of Doa10 with RING domain in MSP1E3D1 | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ERAD-associated E3 ubiquitin-protein ligase DOA10 | | 著者 | Botsch, J.J, Braeuning, B, Schulman, B.A. | | 登録日 | 2023-06-12 | | 公開日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Doa10/MARCH6 architecture interconnects E3 ligase activity with lipid-binding transmembrane channel to regulate SQLE.
Nat Commun, 15, 2024
|
|
8PD0
 
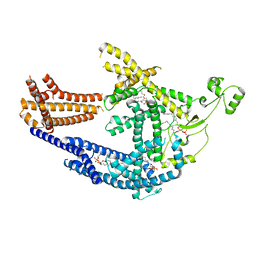 | | cryo-EM structure of Doa10 in MSP1E3D1 | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ERAD-associated E3 ubiquitin-protein ligase DOA10 | | 著者 | Botsch, J.J, Braeuning, B, Schulman, B.A. | | 登録日 | 2023-06-11 | | 公開日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Doa10/MARCH6 architecture interconnects E3 ligase activity with lipid-binding transmembrane channel to regulate SQLE.
Nat Commun, 15, 2024
|
|
8PQL
 
 | | K48-linked ubiquitin chain formation with a cullin-RING E3 ligase and Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2-donor UB-acceptor UB-SIL1 peptide | | 分子名称: | 5-azanylpentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | 登録日 | 2023-07-11 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.76 Å) | | 主引用文献 | Mechanism of millisecond Lys48-linked poly-ubiquitin chain formation by cullin-RING ligases.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8Q7R
 
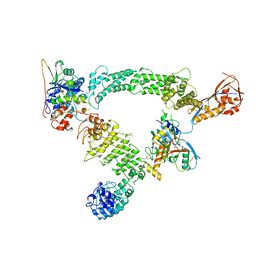 | | Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide | | 分子名称: | 5-azanyl-1-oxidanyl-pentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | 登録日 | 2023-08-16 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.71 Å) | | 主引用文献 | Cullin-RING ligases employ geometrically optimized catalytic partners for substrate targeting.
Mol.Cell, 84, 2024
|
|
8EBM
 
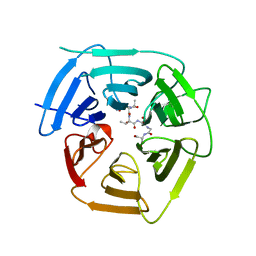 | |
8EBN
 
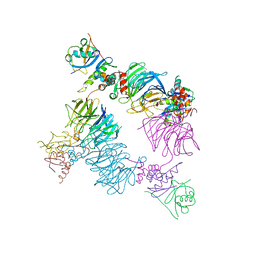 | |
8EBL
 
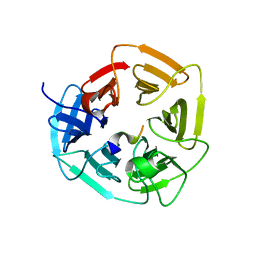 | |
6WY6
 
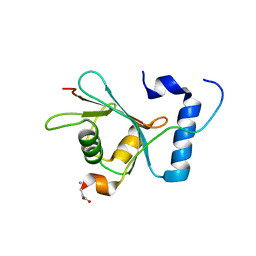 | | Crystal structure of S. cerevisiae Atg8 in complex with Ede1 (1220-1247) | | 分子名称: | Autophagy-related protein 8, EH domain-containing and endocytosis protein 1 | | 著者 | Zheng, Y, Wilfling, F, Baumeister, W, Schulman, B.A. | | 登録日 | 2020-05-12 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.773 Å) | | 主引用文献 | A Selective Autophagy Pathway for Phase-Separated Endocytic Protein Deposits.
Mol.Cell, 80, 2020
|
|
2GJD
 
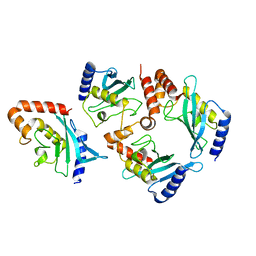 | | Distinct functional domains of Ubc9 dictate cell survival and resistance to genotoxic stress | | 分子名称: | Ubiquitin-conjugating enzyme E2-18 kDa | | 著者 | van Waardenburg, R.C, Duda, D.M, Lancaster, C.S, Schulman, B.A, Bjornsti, M.A. | | 登録日 | 2006-03-30 | | 公開日 | 2006-07-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Distinct functional domains of ubc9 dictate cell survival and resistance to genotoxic stress.
Mol.Cell.Biol., 26, 2006
|
|
6P5W
 
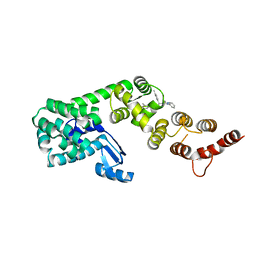 | | Structure of DCN1 bound to 3-methyl-N-((4S,5S)-3-methyl-6-oxo-1-phenyl-4-(p-tolyl)-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)benzamide | | 分子名称: | 3-methyl-N-[(4S,5S)-3-methyl-4-(4-methylphenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]benzamide, Lysozyme,DCN1-like protein 1 chimera | | 著者 | Guy, R.K, Kim, H.S, Hammill, J.T, Scott, D.C, Schulman, B.A. | | 登録日 | 2019-05-31 | | 公開日 | 2019-09-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Discovery of Novel Pyrazolo-pyridone DCN1 Inhibitors Controlling Cullin Neddylation.
J.Med.Chem., 62, 2019
|
|
6P5V
 
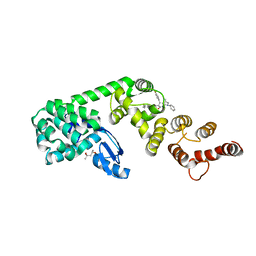 | | Structure of DCN1 bound to N-((4S,5S)-7-ethyl-4-(4-fluorophenyl)-3-methyl-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)-3-methylbenzamide | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lysozyme,DCN1-like protein 1 fusion, N-[(4S,5S)-1-[(1S)-cyclohex-3-en-1-yl]-7-ethyl-4-(4-fluorophenyl)-3-methyl-6-oxo-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]-3-methylbenzamide | | 著者 | Guy, R.K, Kim, H.S, Hammill, J.T, Scott, D.C, Schulman, B.A. | | 登録日 | 2019-05-31 | | 公開日 | 2019-09-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.398 Å) | | 主引用文献 | Discovery of Novel Pyrazolo-pyridone DCN1 Inhibitors Controlling Cullin Neddylation.
J.Med.Chem., 62, 2019
|
|
6SWY
 
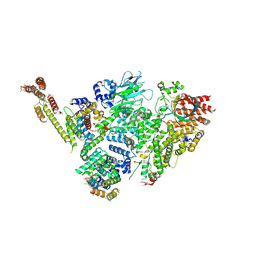 | | Structure of active GID E3 ubiquitin ligase complex minus Gid2 and delta Gid9 RING domain | | 分子名称: | Glucose-induced degradation protein 8, Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10, Vacuolar import and degradation protein 24, ... | | 著者 | Qiao, S, Prabu, J.R, Schulman, B.A. | | 登録日 | 2019-09-24 | | 公開日 | 2019-11-20 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Interconversion between Anticipatory and Active GID E3 Ubiquitin Ligase Conformations via Metabolically Driven Substrate Receptor Assembly
Mol.Cell, 77, 2020
|
|
6TTU
 
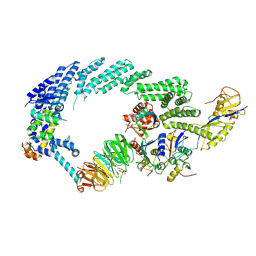 | | Ubiquitin Ligation to substrate by a cullin-RING E3 ligase at 3.7A resolution: NEDD8-CUL1-RBX1 N98R-SKP1-monomeric b-TRCP1dD-IkBa-UB~UBE2D2 | | 分子名称: | CYS-LYS-LYS-ALA-ARG-HIS-ASP-SEP-GLY, Cullin-1, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Baek, K, Prabu, J.R, Schulman, B.A. | | 登録日 | 2019-12-30 | | 公開日 | 2020-02-12 | | 最終更新日 | 2020-03-04 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly.
Nature, 578, 2020
|
|
