2LGX
 
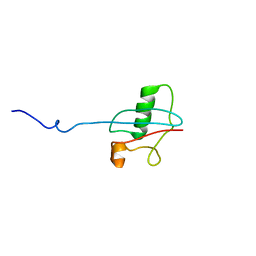 | | NMR structure for Kindle-2 N-terminus | | 分子名称: | Fermitin family homolog 2 | | 著者 | Perera, H.D, Ma, Y, Yang, J, Hirbawi, J, Plow, E.F, Qin, J. | | 登録日 | 2011-08-03 | | 公開日 | 2011-11-30 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Membrane Binding of the N-Terminal Ubiquitin-Like Domain of kindlin-2 Is Crucial for Its Regulation of Integrin Activation.
Structure, 19, 2011
|
|
7D77
 
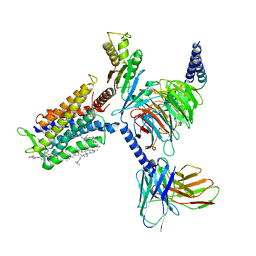 | | Cryo-EM structure of the cortisol-bound adhesion receptor GPR97-Go complex | | 分子名称: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, Adhesion G protein-coupled receptor G3; GPR97, CHOLESTEROL, ... | | 著者 | Ping, Y, Mao, C, Xiao, P, Zhao, R, Jiang, Y, Yang, Z, An, W, Shen, D, Yang, F, Zhang, H, Qu, C, Shen, Q, Tian, C, Li, Z, Li, S, Wang, G, Tao, X, Wen, X, Zhong, Y, Yang, J, Yi, F, Yu, X, Xu, E, Zhang, Y, Sun, J. | | 登録日 | 2020-10-03 | | 公開日 | 2021-02-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structures of the glucocorticoid-bound adhesion receptor GPR97-G o complex.
Nature, 589, 2021
|
|
7D76
 
 | | Cryo-EM structure of the beclomethasone-bound adhesion receptor GPR97-Go complex | | 分子名称: | (8~{S},9~{R},10~{S},11~{S},13~{S},14~{S},16~{S},17~{R})-9-chloranyl-10,13,16-trimethyl-11,17-bis(oxidanyl)-17-(2-oxidanylethanoyl)-6,7,8,11,12,14,15,16-octahydrocyclopenta[a]phenanthren-3-one, Adhesion G protein-coupled receptor G3; GPR97, CHOLESTEROL, ... | | 著者 | Ping, Y, Mao, C, Xiao, P, Zhao, R, Jiang, Y, Yang, Z, An, W, Shen, D, Yang, F, Zhang, H, Qu, C, Shen, Q, Tian, C, Li, Z, Li, S, Wang, G, Tao, X, Wen, X, Zhong, Y, Yang, J, Yi, F, Yu, X, Xu, E, Zhang, Y, Sun, J. | | 登録日 | 2020-10-03 | | 公開日 | 2021-02-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structures of the glucocorticoid-bound adhesion receptor GPR97-G o complex.
Nature, 589, 2021
|
|
2MSU
 
 | |
2YIN
 
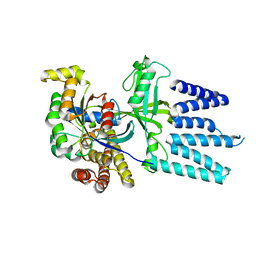 | | STRUCTURE OF THE COMPLEX BETWEEN Dock2 AND Rac1. | | 分子名称: | DEDICATOR OF CYTOKINESIS PROTEIN 2, RAS-RELATED C3 BOTULINUM TOXIN SUBSTRATE 1 | | 著者 | Kulkarni, K.A, Yang, J, Zhang, Z, Barford, D. | | 登録日 | 2011-05-16 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Multiple Factors Confer Specific Cdc42 and Rac Protein Activation by Dedicator of Cytokinesis (Dock) Nucleotide Exchange Factors.
J.Biol.Chem., 286, 2011
|
|
4DFY
 
 | |
6IGB
 
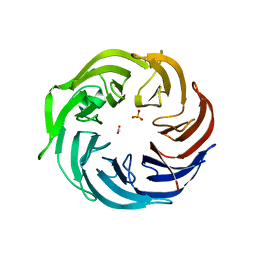 | | the structure of Pseudomonas aeruginosa Periplasmic gluconolactonase, PpgL | | 分子名称: | ACETATE ION, Periplasmic gluconolactonase, PpgL, ... | | 著者 | Song, Y.J, Shen, Y.L, Wang, K.L, Li, T, Zhu, Y.B, Li, C.C, He, L.H, Zhao, N.L, Zhao, C, Yang, J, Huang, Q, Mu, X.Y. | | 登録日 | 2018-09-25 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.651 Å) | | 主引用文献 | Structural and Functional Insights into PpgL, a Metal-Independent beta-Propeller Gluconolactonase That Contributes toPseudomonas aeruginosaVirulence.
Infect.Immun., 87, 2019
|
|
4DG3
 
 | |
5EZY
 
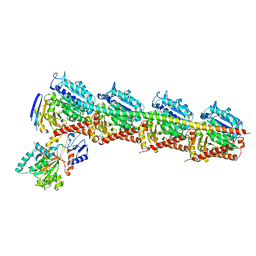 | | Crystal structure of T2R-TTL-taccalonolide AJ complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | 登録日 | 2015-11-27 | | 公開日 | 2017-01-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Mechanism of microtubule stabilization by taccalonolide AJ
Nat Commun, 8, 2017
|
|
5YKJ
 
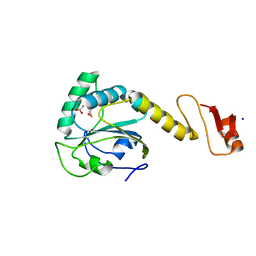 | | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems | | 分子名称: | GLYCEROL, Peroxiredoxin PRX1, mitochondrial, ... | | 著者 | Li, C.C, Yang, J, Yang, M.J, Liu, L, Peng, C.T, Li, T, He, L.H, Song, Y.J, Zhu, Y.B, Zhao, N.L, Zhao, C, Bao, R. | | 登録日 | 2017-10-14 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems
To be published
|
|
5G04
 
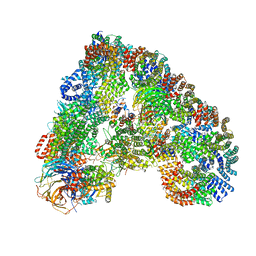 | | Structure of the human APC-Cdc20-Hsl1 complex | | 分子名称: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | 著者 | Zhang, S, Chang, L, Alfieri, C, Zhang, Z, Yang, J, Maslen, S, Skehel, M, Barford, D. | | 登録日 | 2016-03-16 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Molecular Mechanism of Apc/C Activation by Mitotic Phosphorylation.
Nature, 533, 2016
|
|
5H3O
 
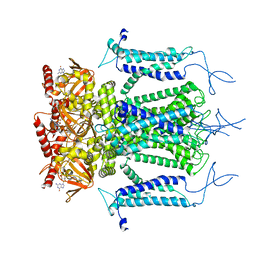 | | Structure of a eukaryotic cyclic nucleotide-gated channel | | 分子名称: | CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel, SODIUM ION | | 著者 | Li, M, Zhou, X, Wang, S, Michailidis, I, Gong, Y, Su, D, Li, H, Li, X, Yang, J. | | 登録日 | 2016-10-26 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of a eukaryotic cyclic-nucleotide-gated channel.
Nature, 542, 2017
|
|
5GJJ
 
 | |
5YKW
 
 | | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems | | 分子名称: | Thioredoxin-3, mitochondrial, peptide THR-PRO-VAL-CYS-THR-THR-GLU-VAL | | 著者 | Li, C.C, Yang, J, Yang, M.J, Liu, L, Peng, C.T, Li, T, He, L.H, Song, Y.J, Zhu, Y.B, Zhao, N.L, Zhao, C, Bao, R. | | 登録日 | 2017-10-16 | | 公開日 | 2018-10-24 | | 最終更新日 | 2019-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems
to be published
|
|
5G05
 
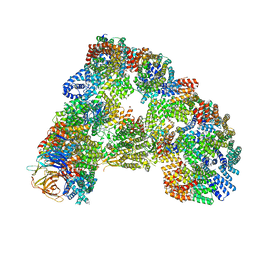 | | Cryo-EM structure of combined apo phosphorylated APC | | 分子名称: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | 著者 | Zhang, S, Chang, L, Alfieri, C, Zhang, Z, Yang, J, Maslen, S, Skehel, M, Barford, D. | | 登録日 | 2016-03-16 | | 公開日 | 2016-05-25 | | 最終更新日 | 2019-09-11 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Molecular Mechanism of Apc/C Activation by Mitotic Phosphorylation.
Nature, 533, 2016
|
|
5H74
 
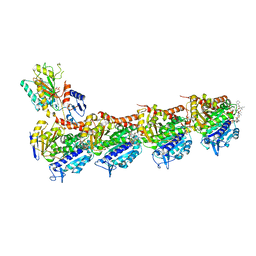 | | Crystal structure of T2R-TTL-14b complex | | 分子名称: | (2~{S},4~{R})-4-[[2-[(1~{R},3~{R})-1-acetyloxy-3-[hexyl-[(2~{S},3~{S})-3-methyl-2-[[(2~{R})-1-methylpiperidin-2-yl]carbonylamino]pentanoyl]amino]-4-methyl-pentyl]-1,3-thiazol-4-yl]carbonylamino]-5-(4-fluorophenyl)-2-methyl-pentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | 登録日 | 2016-11-17 | | 公開日 | 2017-11-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure-Activity Relationship Studies of Tubulysin Analogues: Anticancer N-Alkyltubulysins with Subpicomolar Activity and the Crystal Structure Binding to Tubulin
To Be Published
|
|
4OCZ
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with 1-(1-isobutyrylpiperidin-4-yl)-3-(4-(trifluoromethyl)phenyl)urea | | 分子名称: | 1-[1-(2-methylpropanoyl)piperidin-4-yl]-3-[4-(trifluoromethyl)phenyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | 著者 | Lee, K.S.S, Liu, J, Wagner, K.M, Pakhomova, S, Dong, H, Morriseau, C, Fu, S.H, Yang, J, Wang, P, Ulu, A, Mate, C, Nguyen, L, Wullf, H, Eldin, M.L, Mara, A.A, Newcomer, M.E, Zeldin, D.C, Hammock, B.D. | | 登録日 | 2014-01-09 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy.
J.Med.Chem., 57, 2014
|
|
4OD0
 
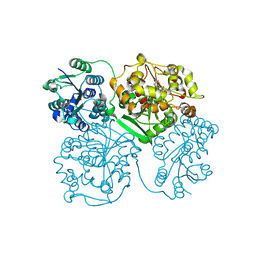 | | Crystal structure of human soluble epoxide hydrolase complexed with 1-(1-propanoylpiperidin-4-yl)-3-[4-(trifluoromethoxy)phenyl]urea | | 分子名称: | 1-(1-propanoylpiperidin-4-yl)-3-[4-(trifluoromethoxy)phenyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | 著者 | Lee, K.S.S, Liu, J, Wagner, K.M, Pakhomova, S, Dong, H, Morisseau, C, Fu, S.H, Yang, J, Wang, P, Ulu, A, Mate, C, Nguyen, L, Wullf, H, Eldin, M.L, Mara, A.A, Newcomer, M.E, Zeldin, D.C, Hammock, B.D. | | 登録日 | 2014-01-09 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy.
J.Med.Chem., 57, 2014
|
|
6JUI
 
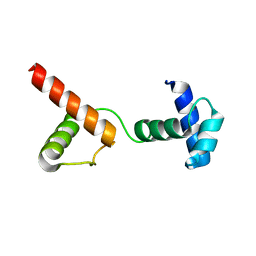 | | The atypical Myb-like protein Cdc5 contains two distinct nucleic acid-binding surfaces | | 分子名称: | Pre-mRNA-splicing factor CEF1 | | 著者 | Wang, C, Li, G, Li, M, Yang, J, Liu, J. | | 登録日 | 2019-04-14 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.402 Å) | | 主引用文献 | Two distinct nucleic acid binding surfaces of Cdc5 regulate development.
Biochem.J., 476, 2019
|
|
6GYS
 
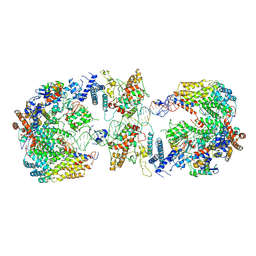 | | Cryo-EM structure of the CBF3-CEN3 complex of the budding yeast kinetochore | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2018-07-01 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6LE5
 
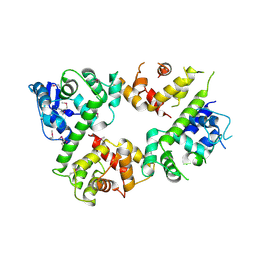 | | Crystal structure of the mitochondrial calcium uptake 1 and 2 heterodimer (MICU1-MICU2 heterodimer) in an apo state | | 分子名称: | Calcium uptake protein 1, mitochondrial, Calcium uptake protein 2 | | 著者 | Park, J, Lee, Y, Park, T, Kang, J.Y, Jin, M, Yang, J, Eom, S.H. | | 登録日 | 2019-11-24 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of the MICU1-MICU2 heterodimer provides insights into the gatekeeping threshold shift.
Iucrj, 7, 2020
|
|
7DIY
 
 | | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-exoribonuclease domain | | 分子名称: | MAGNESIUM ION, ZINC ION, nsp10 protein, ... | | 著者 | Lin, S, Chen, H, Chen, Z.M, Yang, F.L, Ye, F, Zheng, Y, Yang, J, Lin, X, Sun, H.L, Wang, L.L, Wen, A, Cao, Y, Lu, G.W. | | 登録日 | 2020-11-19 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.693 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity.
Nucleic Acids Res., 49, 2021
|
|
2O4X
 
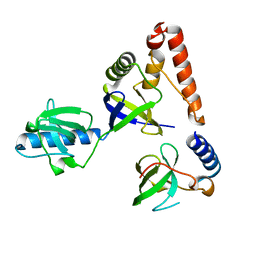 | | Crystal structure of human P100 tudor domain | | 分子名称: | Staphylococcal nuclease domain-containing protein 1 | | 著者 | Shaw, N, Zhao, M, Cheng, C, Xu, H, Yang, J, Silvennoinen, O, Rao, Z, Wang, B.C, Liu, Z.J. | | 登録日 | 2006-12-05 | | 公開日 | 2007-02-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of human P100 tudor domain
To be Published
|
|
6GYU
 
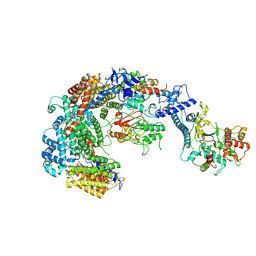 | | Cryo-EM structure of the CBF3-msk complex of the budding yeast kinetochore | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2018-07-02 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6GYP
 
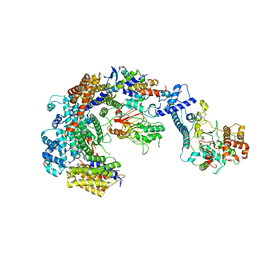 | | Cryo-EM structure of the CBF3-core-Ndc10-DBD complex of the budding yeast kinetochore | | 分子名称: | ARGININE, ASPARAGINE, Centromere DNA-binding protein complex CBF3 subunit A, ... | | 著者 | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2018-07-01 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
