8K92
 
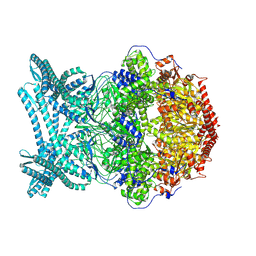 | | CryoEM structure of LonC S582A hexamer with Lysozyme | | Descriptor: | Endopeptidase La, Monothiophosphate | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K8V
 
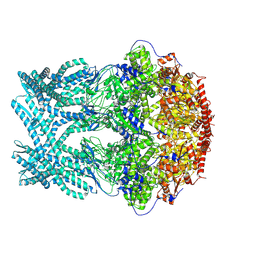 | | CryoEM structure of LonC protease hepatmer, apo state | | Descriptor: | PHOSPHATE ION, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K90
 
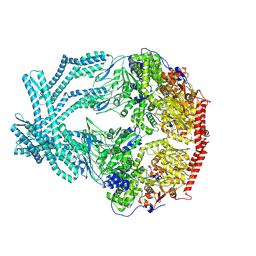 | | CryoEM structure of LonC protease open pentamer in presence of AGS | | Descriptor: | Monothiophosphate, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8XVB
 
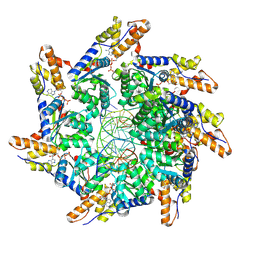 | | Cryo-EM structure of ATP-DNA-MuB filaments | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent target DNA activator B, DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Zhao, X, Zhang, K, Li, S. | | Deposit date: | 2024-01-14 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Elucidating the Architectural dynamics of MuB filaments in bacteriophage Mu DNA transposition
Nat Commun, 15, 2024
|
|
8XVC
 
 | | CryoEM structure of ADP-DNA-MuB conformation1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent target DNA activator B | | Authors: | Zhao, X, Zhang, K, Li, S. | | Deposit date: | 2024-01-14 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Elucidating the Architectural dynamics of MuB filaments in bacteriophage Mu DNA transposition.
Nat Commun, 15, 2024
|
|
8K8Z
 
 | | CryoEM structure of LonC protease hexamer in presence of AGS | | Descriptor: | Monothiophosphate, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K95
 
 | | CryoEM structure of LonC protease open Hexamer, AGS | | Descriptor: | Monothiophosphate, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K8W
 
 | | CryoEM structure of LonC protease open hexamer, apo state | | Descriptor: | endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K91
 
 | | CryoEM structure of LonC S582A hepatmer with Lysozyme | | Descriptor: | Endopeptidase La, Monothiophosphate | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K94
 
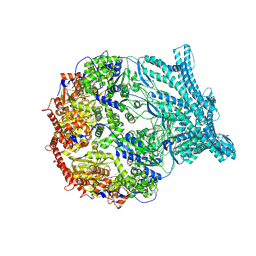 | | CryoEM structure of LonC protease S582A open pentamer with lysozyme | | Descriptor: | Endopeptidase La, Monothiophosphate | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8XVD
 
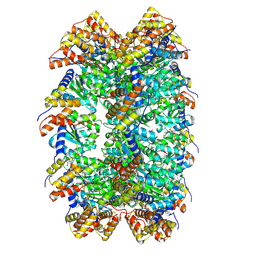 | | CryoEM structure of ADP-DNA-MuB conformation2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent target DNA activator B | | Authors: | Zhao, X, Zhang, K, Li, S. | | Deposit date: | 2024-01-14 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Elucidating the Architectural dynamics of MuB filaments in bacteriophage Mu DNA transposition.
Nat Commun, 15, 2024
|
|
8YTJ
 
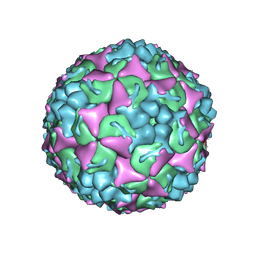 | | Cryo-EM structure of enterovirus A71 mature virion | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2024-03-26 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Cryo-EM structure of enterovirus A71 mature virion in complex with Fab h1A6.2
To Be Published
|
|
8YTB
 
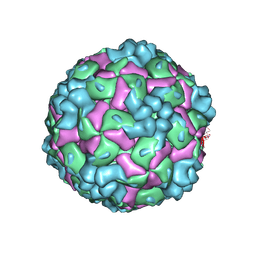 | | Cryo-EM structure of enterovirus A71 empty particle | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2024-03-25 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Cryo-EM structure of enterovirus A71 empty particle
To Be Published
|
|
5J3J
 
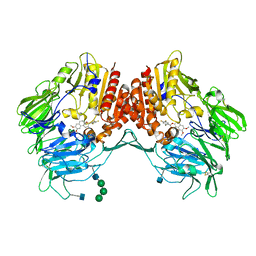 | | Crystal Structure of human DPP-IV in complex with HL1 | | Descriptor: | (2~{S},3~{R})-8,9-dimethoxy-3-[2,4,5-tris(fluoranyl)phenyl]-2,3-dihydro-1~{H}-benzo[f]chromen-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Wu, F, Li, H, Zhao, Z, Zhu, L, Xu, H, Li, S. | | Deposit date: | 2016-03-31 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of human DPP-IV in complex with HL1
To Be Published
|
|
5GZN
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody Heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
5GZO
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
6U9I
 
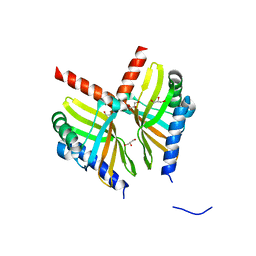 | | Crystal structure of BvnE pinacolase from Penicillium brevicompactum | | Descriptor: | BvnE, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Ye, Y, Du, L, Zhang, X, Newmister, S.A, McCauley, M, Alegre-Requena, J.V, Zhang, W, Mu, S, Minami, A, Fraley, A.E, Adrover-Castellano, M.L, Carney, N, Shende, V.V, Oikawa, H, Kato, H, Tsukamoto, S, Paton, R.S, Williams, R.M, Sherman, D.H, Li, S. | | Deposit date: | 2019-09-09 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.777 Å) | | Cite: | Fungal-derived brevianamide assembly by a stereoselective semipinacolase.
Nat Catal, 3, 2020
|
|
7YHK
 
 | | Cryo-EM structure of the HA trimer of A/Beijing/262/1995(H1N1) in complex with neutralizing antibody 12H5 | | Descriptor: | 12H5 heavy chain, 12H5 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zheng, Q, Li, S, Li, T, Xue, W, Sun, H. | | Deposit date: | 2022-07-13 | | Release date: | 2022-08-17 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses.
Nat Commun, 13, 2022
|
|
4R0S
 
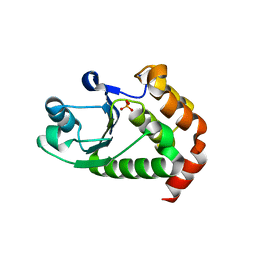 | | Crystal structure of P. aeruginosa TpbA | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein tyrosine phosphatase TpbA | | Authors: | Xu, K, Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2014-08-01 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural and Biochemical Analysis of Tyrosine Phosphatase Related to Biofilm Formation A (TpbA) from the Opportunistic Pathogen Pseudomonas aeruginosa PAO1
PLoS ONE, 10, 2015
|
|
8J60
 
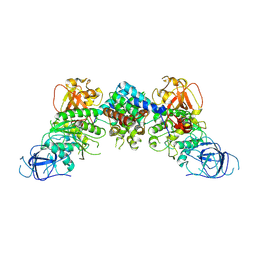 | | Structural and mechanistic insight into ribosomal ITS2 RNA processing by nuclease-kinase machinery | | Descriptor: | LAS1 protein, Polynucleotide 5'-hydroxyl-kinase GRC3 | | Authors: | Chen, J, Chen, H, Li, S, Lin, X, Hu, R, Zhang, K, Liu, L. | | Deposit date: | 2023-04-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural and mechanistic insights into ribosomal ITS2 RNA processing by nuclease-kinase machinery.
Elife, 12, 2024
|
|
8J5Y
 
 | | Structural and mechanistic insight into ribosomal ITS2 RNA processing by nuclease-kinase machinery | | Descriptor: | LAS1 isoform 1, Polynucleotide 5'-hydroxyl-kinase GRC3 | | Authors: | Chen, J, Chen, H, Li, S, Lin, X, Hu, R, Zhang, K, Liu, L. | | Deposit date: | 2023-04-24 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural and mechanistic insights into ribosomal ITS2 RNA processing by nuclease-kinase machinery.
Elife, 12, 2024
|
|
6LGL
 
 | | The atomic structure of varicella-zoster virus A-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Zheng, Q, Li, S. | | Deposit date: | 2019-12-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
6LK5
 
 | | MLKL mutant - T357ES358D | | Descriptor: | Mixed lineage kinase domain-like protein | | Authors: | Wang, H.Y, Li, S, Zhang, Y. | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The MLKL kinase-like domain dimerization is an indispensable step of mammalian MLKL activation in necroptosis signaling.
Cell Death Dis, 12, 2021
|
|
6M54
 
 | | Human apo ferritin frozen on TEM grid with Amorphous nickel titanium alloy supporting film | | Descriptor: | FE (II) ION, Ferritin heavy chain | | Authors: | Huang, X, Zhang, L, Wen, Z, Chen, H, Li, S, Ji, G, Yin, C, Sun, F. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Amorphous nickel titanium alloy film: A new choice for cryo electron microscopy sample preparation.
Prog.Biophys.Mol.Biol., 156, 2020
|
|
1PGV
 
 | | Structural Genomics of Caenorhabditis elegans: tropomodulin C-terminal domain | | Descriptor: | tropomodulin TMD-1 | | Authors: | Symersky, J, Lu, S, Li, S, Chen, L, Meehan, E, Luo, M, Qiu, S, Bunzel, R.J, Luo, D, Arabashi, A, Nagy, L.A, Lin, G, Luan, W.C.-H, Carson, M, Gray, R, Huang, W, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-05-28 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: crystal structure of the tropomodulin C-terminal domain
Proteins, 56, 2004
|
|
