8IFT
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 10 | | Descriptor: | (8S)-N-[(1S)-1-cyano-2-[(3S)-2-oxidanylidenepyrrolidin-3-yl]ethyl]-7-[(2S)-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFQ
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 2 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFS
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 7 | | Descriptor: | (8~{S})-7-[(2~{S})-2-(~{tert}-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-1-cyano-2-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]ethyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFR
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3 | | Descriptor: | (1R,2S,5S)-3-[(2S)-2-(tert-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-N-[(2S)-5-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]hex-3-en-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IGX
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 9 (simnotrelvir, SIM0417, SSD8432) | | Descriptor: | (8~{S})-~{N}-[(1~{S})-1-cyano-2-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]ethyl]-7-[(2~{S})-3,3-dimethyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]butanoyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IGY
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8H59
 
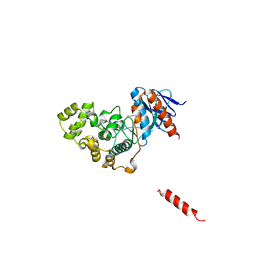 | | A fungal MAP kinase in complex with an inhibitor | | Descriptor: | Mitogen-activated protein kinase MPS1, ~{N}-[(2~{S})-3-(1~{H}-indol-3-yl)-1-(methylamino)-1-oxidanylidene-propan-2-yl]-8-[2-methoxy-5-(trifluoromethyloxy)phenyl]-1,6-naphthyridine-2-carboxamide | | Authors: | Kong, Z, Zhang, X, Wang, D, Liu, J. | | Deposit date: | 2022-10-12 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Aided Identification of an Inhibitor Targets Mps1 for the Management of Plant-Pathogenic Fungi.
Mbio, 14, 2023
|
|
7EZK
 
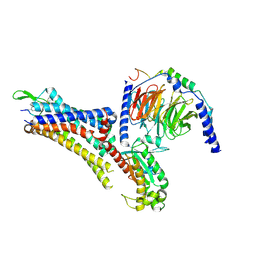 | | Cryo-EM structure of an activated Cholecystokinin A receptor (CCKAR)-Gs complex | | Descriptor: | Chimera of Guanine nucleotide-binding protein G(i) subunit alpha-1 and Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, Cholecystokinin receptor type A, Cholecystokinin-8, ... | | Authors: | Liu, Q.F, Yang, D.H, Zhuang, Y.W, Croll, T.I, Cai, X.Q, Duan, J, Dai, A.T, Yin, W.C, Ye, C.Y, Zhou, F.L, Wu, B.L, Zhao, Q, Xu, H.E, Wang, M.W, Jiang, Y. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ligand recognition and G-protein coupling selectivity of cholecystokinin A receptor
Nat.Chem.Biol., 17, 2021
|
|
7EZM
 
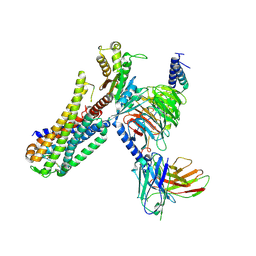 | | Cryo-EM structure of an activated Cholecystokinin A receptor (CCKAR)-Gq complex | | Descriptor: | Cholecystokinin receptor type A, Cholecystokinin-8, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Q.F, Yang, D.H, Zhuang, Y.W, Croll, T.I, Cai, X.Q, Duan, J, Dai, A.T, Yin, W.C, Ye, C.Y, Zhou, F.L, Wu, B.L, Zhao, Q, Xu, H.E, Wang, M.W, Jiang, Y. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ligand recognition and G-protein coupling selectivity of cholecystokinin A receptor
Nat.Chem.Biol., 17, 2021
|
|
7EZH
 
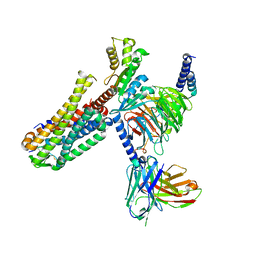 | | Cryo-EM structure of an activated Cholecystokinin A receptor (CCKAR)-Gi complex | | Descriptor: | Cholecystokinin receptor type A, Cholecystokinin-8, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Q.F, Yang, D.H, Zhuang, Y.W, Croll, T.I, Cai, X.Q, Duan, J, Dai, A.T, Yin, W.C, Ye, C.Y, Zhou, F.L, Wu, B.L, Zhao, Q, Xu, H.E, Wang, M.W, Jiang, Y. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ligand recognition and G-protein coupling selectivity of cholecystokinin A receptor
Nat.Chem.Biol., 17, 2021
|
|
6K05
 
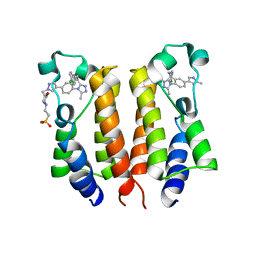 | | Crystal structure of BRD2(BD1)with ligand BY27 bound | | Descriptor: | (6~{R})-~{N}-(4-chlorophenyl)-1-methyl-8-(1-methylpyrazol-4-yl)-5,6-dihydro-4~{H}-[1,2,4]triazolo[4,3-a][1]benzazepin-6-amine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2 | | Authors: | Lu, T, Lu, W, Chen, D, Zhao, Y, Luo, C. | | Deposit date: | 2019-05-05 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Discovery, structural insight, and bioactivities of BY27 as a selective inhibitor of the second bromodomains of BET proteins.
Eur.J.Med.Chem., 182, 2019
|
|
7EPT
 
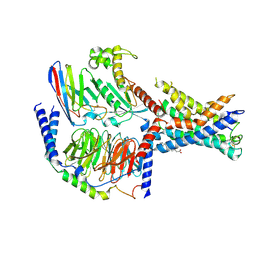 | | Structural basis for the tethered peptide activation of adhesion GPCRs | | Descriptor: | Adhesion G-protein coupled receptor D1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ping, Y.-Q, Xiao, P, Yang, F, Zhao, R.-J, Guo, S.-C, Yan, X, Wu, X, Liebscher, I, Xu, H.E, Sun, J.-P. | | Deposit date: | 2021-04-27 | | Release date: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the tethered peptide activation of adhesion GPCRs.
Nature, 604, 2022
|
|
6K04
 
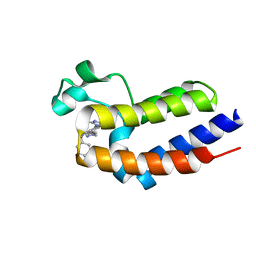 | | Crystal structure of BRD2(BD2)with ligand BY27 bound | | Descriptor: | (6~{R})-~{N}-(4-chlorophenyl)-1-methyl-8-(1-methylpyrazol-4-yl)-5,6-dihydro-4~{H}-[1,2,4]triazolo[4,3-a][1]benzazepin-6-amine, Bromodomain-containing protein 2 | | Authors: | Lu, T, Lu, W, Chen, D, Zhao, Y, Luo, C. | | Deposit date: | 2019-05-05 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Discovery, structural insight, and bioactivities of BY27 as a selective inhibitor of the second bromodomains of BET proteins.
Eur.J.Med.Chem., 182, 2019
|
|
7W4Y
 
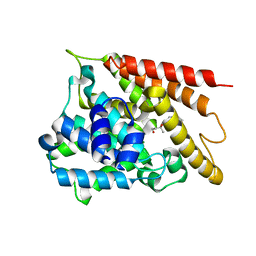 | | Crystal structure of PDE4D catalytic domain complexed with 33a | | Descriptor: | (2R,4S)-6-ethyl-2-(2-hydroxyethyl)-2,8-dimethyl-4-(2-methylprop-1-enyl)-3,4-dihydropyrano[3,2-c][1,8]naphthyridin-5-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2021-11-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.10002947 Å) | | Cite: | Discovery and Structural Optimization of Toddacoumalone Derivatives as Novel PDE4 Inhibitors for the Topical Treatment of Psoriasis.
J.Med.Chem., 65, 2022
|
|
7W4X
 
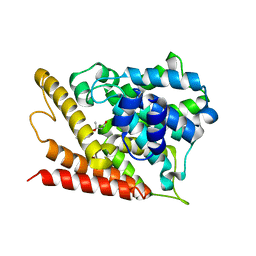 | | Crystal structure of PDE4D catalytic domain complexed with 17 | | Descriptor: | (2R,4S)-2-(2-hydroxyethyl)-2,6-dimethyl-4-(2-methylprop-1-enyl)-3,4-dihydropyrano[3,2-c][1,8]naphthyridin-5-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2021-11-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.20007324 Å) | | Cite: | Discovery and Structural Optimization of Toddacoumalone Derivatives as Novel PDE4 Inhibitors for the Topical Treatment of Psoriasis.
J.Med.Chem., 65, 2022
|
|
7UL1
 
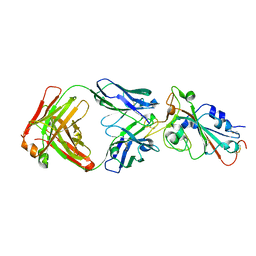 | | Crystal structure of SARS-CoV-2 RBD in complex with the neutralizing IGHV3-53-encoded antibody EH3 isolated from a nonvaccinated pediatric patient | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of EH3, Light chain of EH3, ... | | Authors: | Chen, Y, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2022-04-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD.
Iscience, 26, 2023
|
|
7UL0
 
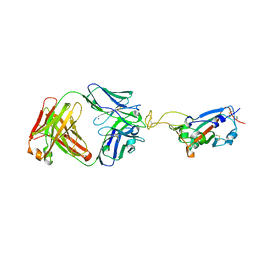 | | Crystal structure of SARS-CoV-2 RBD in complex with the ridge-binding nAb EH8 isolated from a nonvaccinated pediatric patient | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of EH8, ... | | Authors: | Chen, Y, Tolbert, W.D, Bai, X, Pazgier, M. | | Deposit date: | 2022-04-03 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD.
Iscience, 26, 2023
|
|
7EQ1
 
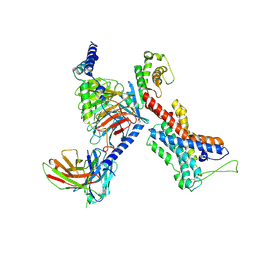 | | GPR114-Gs-scFv16 complex | | Descriptor: | Adhesion G-protein coupled receptor G5, Gs protein alpha subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ping, Y. | | Deposit date: | 2021-04-28 | | Release date: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the tethered peptide activation of adhesion GPCRs.
Nature, 604, 2022
|
|
7X5H
 
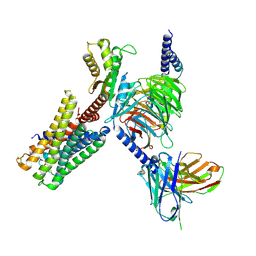 | | Serotonin 5A (5-HT5A) receptor-Gi protein complex | | Descriptor: | 3-(2-azanylethyl)-1H-indole-5-carboxamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tan, Y, Xu, P, Huang, S, Xu, H.E, Jiang, Y. | | Deposit date: | 2022-03-04 | | Release date: | 2022-09-14 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the ligand binding and G i coupling of serotonin receptor 5-HT 5A .
Cell Discov, 8, 2022
|
|
8GZ1
 
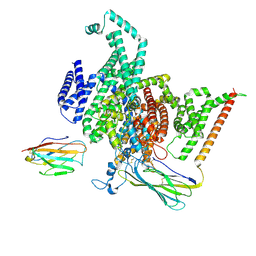 | | Cryo-EM structure of human NaV1.6/beta1/beta2,apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, Sodium channel protein type 8 subunit alpha, ... | | Authors: | Li, Y, Jiang, D. | | Deposit date: | 2022-09-24 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of human Na V 1.6 channel reveals Na + selectivity and pore blockade by 4,9-anhydro-tetrodotoxin.
Nat Commun, 14, 2023
|
|
8GZ2
 
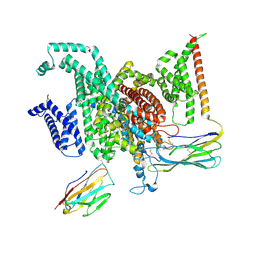 | | Cryo-EM structure of human NaV1.6/beta1/beta2-4,9-anhydro-tetrodotoxin | | Descriptor: | (1R,2S,3S,4R,5R,9S,11S,12S,14R)-7-amino-2,4,12-trihydroxy-2-(hydroxymethyl)-10,13,15-trioxa-6,8-diazapentacyclo[7.4.1.1~3,12~.0~5,11~.0~5,14~]pentadec-7-en-8-ium (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 8 subunit alpha, ... | | Authors: | Li, Y, Jiang, D. | | Deposit date: | 2022-09-24 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human Na V 1.6 channel reveals Na + selectivity and pore blockade by 4,9-anhydro-tetrodotoxin.
Nat Commun, 14, 2023
|
|
8HKK
 
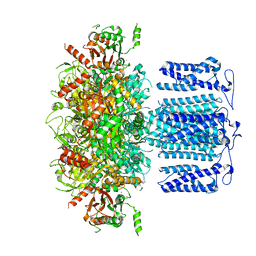 | | ion channel | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, SODIUM ION, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-27 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
8HKQ
 
 | | ion channel | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, SODIUM ION, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-27 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
8HKF
 
 | | ion channel | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, ZINC ION | | Authors: | Jiang, D.H, Zhang, Z.T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
8HKM
 
 | | ion channel | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, ZINC ION, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-27 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
