7JW7
 
 | | Structure of monobody 27 human MLKL pseudokinase domain complex | | Descriptor: | Mixed lineage kinase domain-like protein, Monobody 27 | | Authors: | Meng, Y, Garnish, S.E, Koide, A, Koide, S, Czabotar, P.E, Murphy, J.M. | | Deposit date: | 2020-08-25 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Conformational interconversion of MLKL and disengagement from RIPK3 precede cell death by necroptosis.
Nat Commun, 12, 2021
|
|
7JXU
 
 | | Structure of monobody 32 human MLKL pseudokinase domain complex | | Descriptor: | 1,2-ETHANEDIOL, Mixed lineage kinase domain-like protein, Monobody 32 | | Authors: | Meng, Y, Garnish, S.E, Koide, A, Koide, S, Czabotar, P.E, Murphy, J.M. | | Deposit date: | 2020-08-28 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Conformational interconversion of MLKL and disengagement from RIPK3 precede cell death by necroptosis.
Nat Commun, 12, 2021
|
|
7K02
 
 | |
7LK3
 
 | | Crystal structure of untwinned human GABARAPL2 | | Descriptor: | 1,2-ETHANEDIOL, Gamma-aminobutyric acid receptor-associated protein-like 2 | | Authors: | Scicluna, K, Dewson, G, Czabotar, P.E, Birkinshaw, R.W. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new crystal form of GABARAPL2.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
8SLZ
 
 | |
2VOI
 
 | | Structure of mouse A1 bound to the Bid BH3-domain | | Descriptor: | BCL-2-RELATED PROTEIN A1, BH3-INTERACTING DOMAIN DEATH AGONIST P13, CHLORIDE ION | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
2VOG
 
 | | Structure of mouse A1 bound to the Bmf BH3-domain | | Descriptor: | BCL-2-MODIFYING FACTOR, BCL-2-RELATED PROTEIN A1, CHLORIDE ION | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
2VOH
 
 | | Structure of mouse A1 bound to the Bak BH3-domain | | Descriptor: | BCL-2 HOMOLOGOUS ANTAGONIST/KILLER, BCL-2-RELATED PROTEIN A1, CITRIC ACID, ... | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
2VOF
 
 | | Structure of mouse A1 bound to the Puma BH3-domain | | Descriptor: | BCL-2-BINDING COMPONENT 3, BCL-2-RELATED PROTEIN A1, CHLORIDE ION | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
2VTY
 
 | | Vaccinia virus anti-apoptotic F1L is a novel Bcl-2-like domain swapped dimer | | Descriptor: | PROTEIN F1 | | Authors: | Kvansakul, M, Yang, H, Fairlie, W.D, Czabotar, P.E, Fischer, S.F, Perugini, M.A, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2008-05-19 | | Release date: | 2008-06-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Vaccinia Virus Anti-Apoptotic F1L is a Novel Bcl-2-Like Domain-Swapped Dimer that Binds a Highly Selective Subset of Bh3-Containing Death Ligands.
Cell Death Differ., 15, 2008
|
|
6MCY
 
 | | Crystal structure of mouse Bak | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2 homologous antagonist/killer, FORMIC ACID | | Authors: | Brouwer, J.M, Czabotar, P.E, Colman, P.M. | | Deposit date: | 2018-09-03 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | A small molecule interacts with VDAC2 to block mouse BAK-driven apoptosis.
Nat.Chem.Biol., 15, 2019
|
|
6O0L
 
 | | crystal structure of BCL-2 G101V mutation with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, CHLORIDE ION, ... | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-16 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
6O0P
 
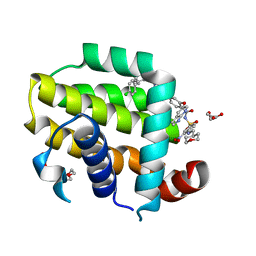 | | crystal structure of BCL-2 G101A mutation with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, DI(HYDROXYETHYL)ETHER | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-17 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
6O0K
 
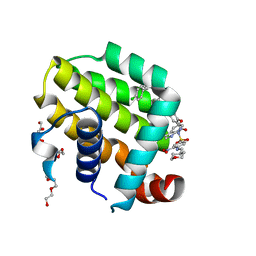 | | crystal structure of BCL-2 with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2, NONAETHYLENE GLYCOL | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-16 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
6O0O
 
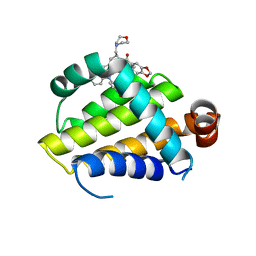 | | crystal structure of BCL-2 G101V mutation with S55746 | | Descriptor: | Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, ~{N}-(4-hydroxyphenyl)-3-[6-[[(3~{S})-3-(morpholin-4-ylmethyl)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]-1,3-benzodioxol-5-yl]-~{N}-phenyl-5,6,7,8-tetrahydroindolizine-1-carboxamide | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-17 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
6O0M
 
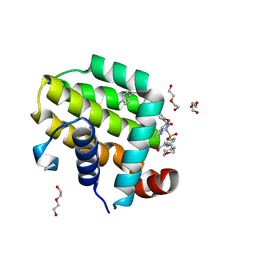 | | crystal structure of BCL-2 F104L mutation with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, DI(HYDROXYETHYL)ETHER | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-16 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
6N64
 
 | | Crystal structure of mouse SMCHD1 hinge domain | | Descriptor: | Structural maintenance of chromosomes flexible hinge domain-containing protein 1, Uncharacterized peptide from Structural maintenance of chromosomes flexible hinge domain-containing protein 1 | | Authors: | Birkinshaw, R.W, Chen, K, Czabotar, P.E, Blewitt, M.E, Murphy, J.M. | | Deposit date: | 2018-11-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the hinge domain of Smchd1 reveals its dimerization mode and nucleic acid-binding residues.
Sci.Signal., 13, 2020
|
|
6O5Z
 
 | | Crystal Structure of the human MLKL pseudokinase domain bound to compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-fluoranyl-5-(trifluoromethyl)phenyl]-3-[4-[methyl-[2-[(3-sulfamoylphenyl)amino]pyrimidin-4-yl]amino]phenyl]urea, Mixed lineage kinase domain-like protein | | Authors: | Cowan, A.D, Murphy, J.M, Pierotti, C.L, Lessene, G.L, Czabotar, P.E. | | Deposit date: | 2019-03-04 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Potent Inhibition of Necroptosis by Simultaneously Targeting Multiple Effectors of the Pathway.
Acs Chem.Biol., 15, 2020
|
|
5FMK
 
 | | Bcl-xL with Bak BH3 complex | | Descriptor: | BCL-2 HOMOLOGOUS ANTAGONIST/KILLER, BCL-XL, GLYCEROL | | Authors: | Fairlie, W.D, Lee, E.F, Smith, B.J, Czabotar, P.E, Colman, P.M. | | Deposit date: | 2015-11-06 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.731 Å) | | Cite: | Physiological Restraint of Bak by Bcl-Xl is Essential for Cell Survival.
Genes Dev., 30, 2016
|
|
4ZIE
 
 | | Crystal Structure of core/latch dimer of Bax in complex with BimBH3 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX, Bcl-2-like protein 11 | | Authors: | Krishna Kumar, K, Robin, A.Y, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
4ZIF
 
 | | Crystal Structure of core/latch dimer of Bax in complex with BimBH3mini | | Descriptor: | Apoptosis regulator BAX, Bcl-2-like protein 11 | | Authors: | Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
4ZIG
 
 | | Crystal Structure of core/latch dimer of Bax in complex with BidBH3mini | | Descriptor: | Apoptosis regulator BAX, BH3-interacting domain death agonist | | Authors: | Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
4ZIH
 
 | | Crystal Structure of core/latch dimer of Bax in complex with BimBH3mini | | Descriptor: | Apoptosis regulator BAX, Bcl-2-like protein 11 | | Authors: | Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
6UXO
 
 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with DDM | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Bcl-2 homologous antagonist/killer, ... | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-11-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UXQ
 
 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with POPC and C8E4 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, ... | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-11-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
