5KDR
 
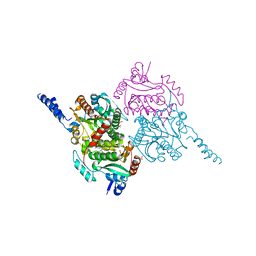 | | The crystal structure of carboxyltransferase from Staphylococcus Aureus bound to the antimicrobial agent moiramide B. | | Descriptor: | 1,2-ETHANEDIOL, Acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, Acetyl-coenzyme A carboxylase carboxyl transferase subunit beta, ... | | Authors: | Silvers, M.A, Pakhomova, S, Neau, D, Silvers, W.C, Anzalone, N, Taylor, C.M, Waldrop, G.L. | | Deposit date: | 2016-06-08 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Carboxyltransferase from Staphylococcus aureus Bound to the Antibacterial Agent Moiramide B.
Biochemistry, 55, 2016
|
|
5UYT
 
 | | Crystal structure of ice binding protein from an Antarctic bacterium Flavobacteriaceae | | Descriptor: | Ice-binding protein, NITRATE ION | | Authors: | Wang, C, Pakhomova, S, Newcomer, M.E, Christner, B.C, Luo, B.-H. | | Deposit date: | 2017-02-24 | | Release date: | 2017-10-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of antifreeze activity of a bacterial multi-domain antifreeze protein.
PLoS ONE, 12, 2017
|
|
2P7M
 
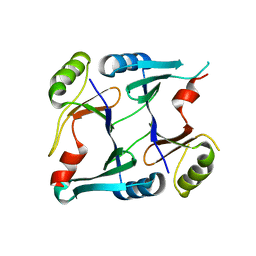 | | Crystal structure of monoclinic form of genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes at pH 6.5 | | Descriptor: | Glyoxalase family protein | | Authors: | Fillgrove, K.L, Pakhomova, S, Schaab, M, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2007-03-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria monocytogenes.
Biochemistry, 46, 2007
|
|
2P7L
 
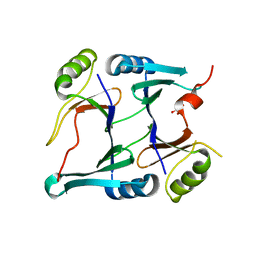 | | Crystal structure of monoclinic form of genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes at pH 5.75 | | Descriptor: | GLYCEROL, Glyoxalase family protein | | Authors: | Fillgrove, K.L, Pakhomova, S, Schaab, M, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2007-03-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria monocytogenes.
Biochemistry, 46, 2007
|
|
2P7P
 
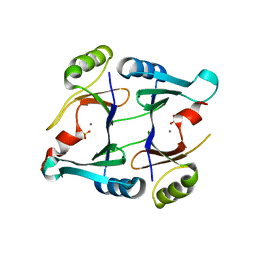 | | Crystal structure of genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes complexed with MN(II) and sulfate ion | | Descriptor: | Glyoxalase family protein, MANGANESE (II) ION, SULFATE ION | | Authors: | Fillgrove, K.L, Pakhomova, S, Schaab, M, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2007-03-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria monocytogenes.
Biochemistry, 46, 2007
|
|
2AZT
 
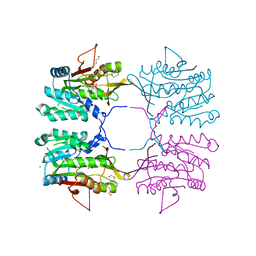 | | Crystal structure of H176N mutant of human Glycine N-Methyltransferase | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Luka, Z, Pakhomova, S, Luka, Y, Newcomer, M.E, Wagner, C. | | Deposit date: | 2005-09-12 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Destabilization of human glycine N-methyltransferase by H176N mutation.
Protein Sci., 16, 2007
|
|
4HAI
 
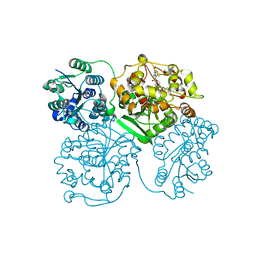 | | Crystal structure of human soluble epoxide hydrolase complexed with N-cycloheptyl-1-(mesitylsulfonyl)piperidine-4-carboxamide. | | Descriptor: | Bifunctional epoxide hydrolase 2, MAGNESIUM ION, N-cycloheptyl-1-[(2,4,6-trimethylphenyl)sulfonyl]piperidine-4-carboxamide, ... | | Authors: | Pecic, S, Pakhomova, S, Newcomer, M.E, Morisseau, C, Hammock, B.D, Zhu, Z, Deng, S. | | Deposit date: | 2012-09-26 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Synthesis and structure-activity relationship of piperidine-derived non-urea soluble epoxide hydrolase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6OI9
 
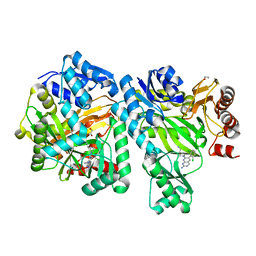 | | Crystal Structure of E. coli Biotin Carboxylase Complexed with 7-[3-(aminomethyl)pyrrolidin-1-yl]-6-(2,6-dichlorophenyl)pyrido[2,3-d]pyrimidin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 7-[(3S)-3-(aminomethyl)pyrrolidin-1-yl]-6-(2,6-dichlorophenyl)pyrido[2,3-d]pyrimidin-2-amine, Biotin carboxylase | | Authors: | Andrews, L.D, Kane, T.R, Dozzo, P, Haglund, C.M, Hilderbrandt, D.J, Linsell, M.S, Machajewski, T, McEnroe, G, Serio, A.W, Wlasichuk, K.B, Neau, D.B, Pakhomova, S, Waldrop, G.L, Sharp, M, Pogliano, J, Cirz, R, Cohen, F. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Optimization and Mechanistic Characterization of Pyridopyrimidine Inhibitors of Bacterial Biotin Carboxylase.
J.Med.Chem., 62, 2019
|
|
6OJH
 
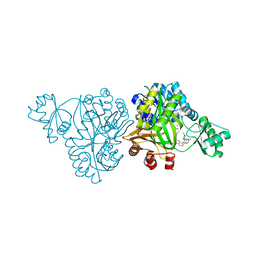 | | Crystal Structure of Haemophilus Influenzae Biotin Carboxylase Complexed with (R)-7-(3-aminopyrrolidin-1-yl)-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine | | Descriptor: | 7-[(3R)-3-aminopyrrolidin-1-yl]-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine, ACETATE ION, Biotin carboxylase, ... | | Authors: | Andrews, L.D, Kane, T.R, Dozzo, P, Haglund, C.M, Hilderbrandt, D.J, Linsell, M.S, Machajewski, T, McEnroe, G, Serio, A.W, Wlasichuk, K.B, Neau, D.B, Pakhomova, S, Waldrop, G.L, Sharp, M, Pogliano, J, Cirz, R, Cohen, F. | | Deposit date: | 2019-04-11 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Haemophilus Influenzae Biotin Carboxylase Complexed with (R)-7-(3-aminopyrrolidin-1-yl)-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine
To Be Published
|
|
4J03
 
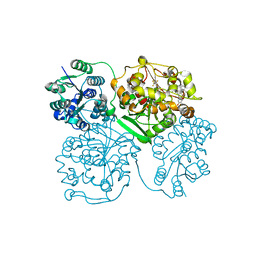 | | Crystal structure of human soluble epoxide hydrolase complexed with fulvestrant | | Descriptor: | (7beta,9beta,13alpha,17beta)-7-{9-[(R)-(4,4,5,5,5-pentafluoropentyl)sulfinyl]nonyl}estra-1(10),2,4-triene-3,17-diol, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Morisseau, C, Pakhomova, S, Hwang, S.H, Newcomer, M.E, Hammock, B.D. | | Deposit date: | 2013-01-30 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Inhibition of soluble epoxide hydrolase by fulvestrant and sulfoxides.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6OI8
 
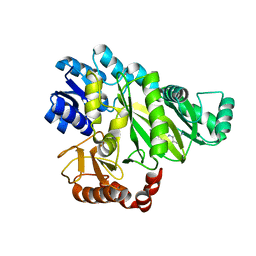 | | Crystal Structure of Haemophilus Influenzae Biotin Carboxylase Complexed with 7-((1R,5S,6s)-6-amino-3-azabicyclo[3.1.0]hexan-3-yl)-6-(2-chloro-6-(pyridin-3-yl)phenyl)pyrido[2,3-d]pyrimidin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 7-[(1R,5S,6s)-6-amino-3-azabicyclo[3.1.0]hexan-3-yl]-6-[2-chloro-6-(pyridin-3-yl)phenyl]pyrido[2,3-d]pyrimidin-2-amine, Biotin carboxylase | | Authors: | Andrews, L.D, Kane, T.R, Dozzo, P, Haglund, C.M, Hilderbrandt, D.J, Linsell, M.S, Machajewski, T, McEnroe, G, Serio, A.W, Wlasichuk, K.B, Neau, D.B, Pakhomova, S, Waldrop, G.L, Sharp, M, Pogliano, J, Cirz, R, Cohen, F. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Optimization and Mechanistic Characterization of Pyridopyrimidine Inhibitors of Bacterial Biotin Carboxylase.
J.Med.Chem., 62, 2019
|
|
4KUM
 
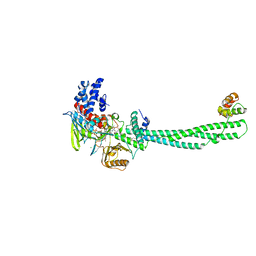 | | Structure of LSD1-CoREST-Tetrahydrofolate complex | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Luka, Z, Pakhomova, S, Loukachevitch, L.V, Calcutt, M.W, Newcomer, M.E, Wagner, C. | | Deposit date: | 2013-05-22 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of the histone lysine specific demethylase LSD1 complexed with tetrahydrofolate.
Protein Sci., 23, 2014
|
|
4HR7
 
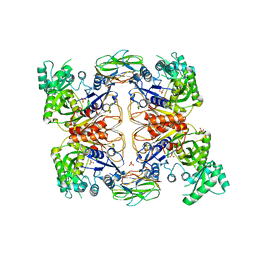 | | Crystal Structure of Biotin Carboxyl Carrier Protein-Biotin Carboxylase Complex from E.coli | | Descriptor: | 1,2-ETHANEDIOL, Biotin carboxyl carrier protein of acetyl-CoA carboxylase, Biotin carboxylase, ... | | Authors: | Broussard, T.C, Kobe, M.J, Pakhomova, S, Neau, D.B, Price, A.E, Champion, T.S, Waldrop, G.L. | | Deposit date: | 2012-10-26 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | The three-dimensional structure of the biotin carboxylase-biotin carboxyl carrier protein complex of E. coli acetyl-CoA carboxylase.
Structure, 21, 2013
|
|
3THS
 
 | | Crystal structure of rat native liver Glycine N-methyltransferase complexed with 5-methyltetrahydrofolate pentaglutamate | | Descriptor: | 5-methyltetrahydrofolate pentaglutamate, BETA-MERCAPTOETHANOL, Glycine N-methyltransferase, ... | | Authors: | Luka, Z, Pakhomova, S, Loukachevitch, L.V, Newcomer, M.E, Wagner, C. | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Differences in folate-protein interactions result in differing inhibition of native rat liver and recombinant glycine N-methyltransferase by 5-methyltetrahydrofolate.
Biochim.Biophys.Acta, 1824, 2011
|
|
3THR
 
 | | Crystal structure of rat native liver Glycine N-methyltransferase complexed with 5-methyltetrahydrofolate monoglutamate | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, Glycine N-methyltransferase, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Luka, Z, Pakhomova, S, Loukachevitch, L.V, Newcomer, M.E, Wagner, C. | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differences in folate-protein interactions result in differing inhibition of native rat liver and recombinant glycine N-methyltransferase by 5-methyltetrahydrofolate.
Biochim.Biophys.Acta, 1824, 2011
|
|
2IDK
 
 | | Crystal Structure of Rat Glycine N-Methyltransferase Complexed With Folate | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, Glycine N-methyltransferase | | Authors: | Luka, Z, Pakhomova, S, Loukachevitch, L.V, Egli, M, Newcomer, M.E, Wagner, C. | | Deposit date: | 2006-09-15 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 5-methyltetrahydrofolate is bound in intersubunit areas of rat liver folate-binding protein glycine N-methyltransferase.
J.Biol.Chem., 282, 2007
|
|
2IDJ
 
 | | Crystal Structure of Rat Glycine N-Methyltransferase Apoprotein, Monoclinic Form | | Descriptor: | CALCIUM ION, Glycine N-methyltransferase | | Authors: | Luka, Z, Pakhomova, S, Loukachevitch, L.V, Egli, M, Newcomer, M.E, Wagner, C. | | Deposit date: | 2006-09-15 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 5-methyltetrahydrofolate is bound in intersubunit areas of rat liver folate-binding protein glycine N-methyltransferase.
J.Biol.Chem., 282, 2007
|
|
4RZQ
 
 | | Structural Analysis of Substrate, Reaction Intermediate and Product Binding in Haemophilus influenzae Biotin Carboxylase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Biotin carboxylase, methyl (3aS,4S,6aR)-4-(5-methoxy-5-oxopentyl)-2-oxohexahydro-1H-thieno[3,4-d]imidazole-1-carboxylate | | Authors: | Broussard, T.C, Pakhomova, S, Neau, D.B, Champion, T.S, Bonnot, R, Waldrop, G.L. | | Deposit date: | 2014-12-23 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Analysis of Substrate, Reaction Intermediate, and Product Binding in Haemophilus influenzae Biotin Carboxylase.
Biochemistry, 54, 2015
|
|
2P7O
 
 | | Crystal structure of genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes (tetragonal form) | | Descriptor: | Glyoxalase family protein, MANGANESE (II) ION | | Authors: | Fillgrove, K.L, Pakhomova, S, Schaab, M, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2007-03-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria monocytogenes.
Biochemistry, 46, 2007
|
|
2P7K
 
 | | Crystal structure of genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes (hexagonal form) | | Descriptor: | CITRIC ACID, Glyoxalase family protein | | Authors: | Fillgrove, K.L, Pakhomova, S, Schaab, M, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2007-03-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria monocytogenes.
Biochemistry, 46, 2007
|
|
2P7Q
 
 | | Crystal structure of E126Q mutant of genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes complexed with MN(II) and 1S,2S-dihydroxypropylphosphonic acid | | Descriptor: | Glyoxalase family protein, MANGANESE (II) ION, [(1S,2S)-1,2-DIHYDROXYPROPYL]PHOSPHONIC ACID | | Authors: | Fillgrove, K.L, Pakhomova, S, Schaab, M, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2007-03-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria monocytogenes.
Biochemistry, 46, 2007
|
|
1R9C
 
 | | Crystal Structure of Fosfomycin Resistance Protein FosX from Mesorhizobium Loti | | Descriptor: | MANGANESE (II) ION, glutathione transferase | | Authors: | Fillgrove, K.L, Pakhomova, S, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2003-10-28 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Mechanistic diversity of fosfomycin resistance in pathogenic microorganisms.
J.Am.Chem.Soc., 125, 2003
|
|
4OCZ
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with 1-(1-isobutyrylpiperidin-4-yl)-3-(4-(trifluoromethyl)phenyl)urea | | Descriptor: | 1-[1-(2-methylpropanoyl)piperidin-4-yl]-3-[4-(trifluoromethyl)phenyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Lee, K.S.S, Liu, J, Wagner, K.M, Pakhomova, S, Dong, H, Morriseau, C, Fu, S.H, Yang, J, Wang, P, Ulu, A, Mate, C, Nguyen, L, Wullf, H, Eldin, M.L, Mara, A.A, Newcomer, M.E, Zeldin, D.C, Hammock, B.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy.
J.Med.Chem., 57, 2014
|
|
4PAA
 
 | | Crystal structure of the mature form of rat DMGDH complexed with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, Dimethylglycine dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Luka, Z, Pakhomova, S, Loukachevitch, L.V, Newcomer, M.E, Wagner, C. | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Folate in demethylation: The crystal structure of the rat dimethylglycine dehydrogenase complexed with tetrahydrofolate.
Biochem.Biophys.Res.Commun., 449, 2014
|
|
4MV4
 
 | | Crystal Structure of Biotin Carboxylase from Haemophilus influenzae in Complex with AMPPCP and Mg2 | | Descriptor: | 1,2-ETHANEDIOL, Biotin carboxylase, CHLORIDE ION, ... | | Authors: | Broussard, T.C, Pakhomova, S, Neau, D.B, Champion, T.S, Bonnot, R.J, Waldrop, G.L. | | Deposit date: | 2013-09-23 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural Analysis of Substrate, Reaction Intermediate, and Product Binding in Haemophilus influenzae Biotin Carboxylase.
Biochemistry, 54, 2015
|
|
