1SHW
 
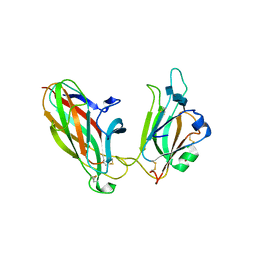 | | EphB2 / EphrinA5 Complex Structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin type-B receptor 2, Ephrin-A5, ... | | Authors: | Himanen, J.P, Chumley, M.J, Lackmann, M, Li, C, Barton, W.A, Jeffrey, P.D, Vearing, C, Geleick, D, Feldheim, D.A, Boyd, A.W. | | Deposit date: | 2004-02-26 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Repelling class discrimination: ephrin-A5 binds to and activates EphB2 receptor signaling
Nat.Neurosci., 7, 2004
|
|
3IYS
 
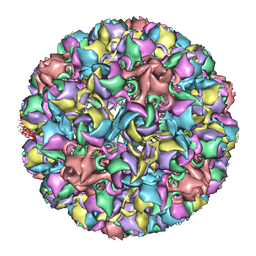 | | Homology model of avian polyomavirus asymmetric unit | | Descriptor: | Major capsid protein VP1 | | Authors: | Shen, P.S, Enderlein, D, Nelson, C.D.S, Carter, W.S, Kawano, M, Xing, L, Swenson, R.D, Olson, N.H, Baker, T.S, Cheng, R.H, Atwood, W.J, Johne, R, Belnap, D.M. | | Deposit date: | 2010-04-19 | | Release date: | 2011-01-26 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.3 Å) | | Cite: | The structure of avian polyomavirus reveals variably sized capsids, non-conserved inter-capsomere interactions, and a possible location of the minor capsid protein VP4.
Virology, 411, 2011
|
|
4AP5
 
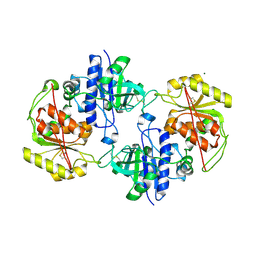 | | Crystal structure of human POFUT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GDP-FUCOSE PROTEIN O-FUCOSYLTRANSFERASE 2, ... | | Authors: | Chen, C, Keusch, J.J, Klein, D, Hess, D, Hofsteenge, J, Gut, H. | | Deposit date: | 2012-03-30 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structure of Human Pofut2: Insights Into Thrombospondin Type 1 Repeat Fold and O-Fucosylation.
Embo J., 31, 2012
|
|
4AP6
 
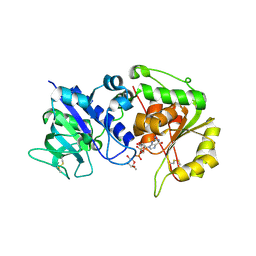 | | Crystal structure of human POFUT2 E54A mutant in complex with GDP- fucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GDP-FUCOSE PROTEIN O-FUCOSYLTRANSFERASE 2, ... | | Authors: | Chen, C, Keusch, J.J, Klein, D, Hess, D, Hofsteenge, J, Gut, H. | | Deposit date: | 2012-03-30 | | Release date: | 2012-08-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Structure of Human Pofut2: Insights Into Thrombospondin Type 1 Repeat Fold and O-Fucosylation.
Embo J., 31, 2012
|
|
4W7T
 
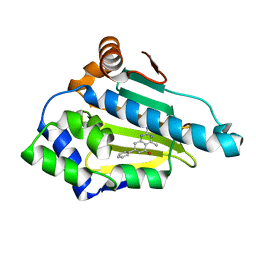 | | Crystal Structure of Hsp90-alpha N-domain Bound to the Inhibitor NVP-HSP990 | | Descriptor: | (7S)-2-amino-4-methyl-7-phenyl-7,8-dihydroquinazolin-5(6H)-one, Heat shock protein HSP 90-alpha | | Authors: | Bellamacina, C.R, Shafer, C.M, Bussiere, D. | | Deposit date: | 2014-08-22 | | Release date: | 2014-11-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Structure-Activity Relationship, and in Vivo Characterization of the Development Candidate NVP-HSP990.
J.Med.Chem., 57, 2014
|
|
8GK7
 
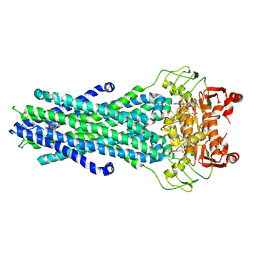 | | MsbA bound to cerastecin C | | Descriptor: | 2-[(4-butylbenzene-1-sulfonyl)amino]-5-[(3-{4-[(4-butylbenzene-1-sulfonyl)amino]-3-carboxyanilino}-3-oxopropyl)carbamoyl]benzoic acid, Lipid A export ATP-binding/permease protein MsbA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Chen, Y, Klein, D. | | Deposit date: | 2023-03-17 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cerastecins inhibit membrane lipooligosaccharide transport in drug-resistant Acinetobacter baumannii.
Nat Microbiol, 9, 2024
|
|
8IDE
 
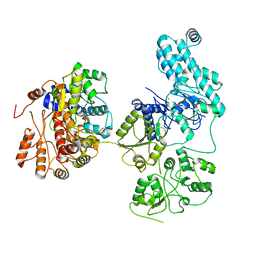 | | Structure of an ancient TsaD-TsaC-SUA5-TcdA modular enzyme (TsaN) | | Descriptor: | MANGANESE (II) ION, N(6)-L-threonylcarbamoyladenine synthase | | Authors: | Zhang, Z.L, Jin, M.Q, Yu, Z.J, Chen, W, Wang, X.L, Lei, D.S, Zhang, W.H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structure-function analysis of an ancient TsaD-TsaC-SUA5-TcdA modular enzyme reveals a prototype of tRNA t6A and ct6A synthetases.
Nucleic Acids Res., 51, 2023
|
|
4U93
 
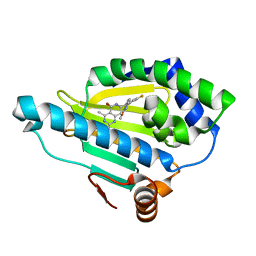 | | Crystal Structure of Hsp90-alpha N-domain Bound to the Inhibitor NVP-HSP990 | | Descriptor: | (7R)-2-amino-7-[4-fluoro-2-(6-methoxypyridin-2-yl)phenyl]-4-methyl-7,8-dihydropyrido[4,3-d]pyrimidin-5(6H)-one, Heat shock protein HSP 90-alpha | | Authors: | Bellamacina, C.R, Shafer, C.M, Bussiere, D. | | Deposit date: | 2014-08-05 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design, Structure-Activity Relationship, and in Vivo Characterization of the Development Candidate NVP-HSP990.
J.Med.Chem., 57, 2014
|
|
4QEI
 
 | | Two distinct conformational states of GlyRS captured in crystal lattice | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glycine--tRNA ligase, tRNA-Gly-CCC-2-2 | | Authors: | Xie, W, Qin, X, Deng, X, Zhang, Q, Li, Q. | | Deposit date: | 2014-05-16 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.875 Å) | | Cite: | Large Conformational Changes of Insertion 3 in Human Glycyl-tRNA Synthetase (hGlyRS) during Catalysis
J.Biol.Chem., 291, 2016
|
|
8WC3
 
 | | Cryo-EM structure of the SEP363856-bound mTAAR1-Gs complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC7
 
 | | Cryo-EM structure of the ZH8667-bound mTAAR1-Gs complex | | Descriptor: | 2-[4-(3-fluorophenyl)phenyl]ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC4
 
 | | Cryo-EM structure of the ZH8651-bound mTAAR1-Gs complex | | Descriptor: | 2-(4-bromophenyl)ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC9
 
 | | Cryo-EM structure of the ZH8651-bound mTAAR1-Gq complex | | Descriptor: | 2-(4-bromophenyl)ethanamine, Engineered G-alpha-q subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC8
 
 | | Cryo-EM structure of the ZH8651-bound hTAAR1-Gs complex | | Descriptor: | 2-(4-bromophenyl)ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC5
 
 | | Cryo-EM structure of the TMA-bound mTAAR1-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCA
 
 | | Cryo-EM structure of the PEA-bound hTAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCB
 
 | | Cryo-EM structure of the CHA-bound mTAAR1-Gq complex | | Descriptor: | CYCLOHEXYLAMMONIUM ION, Engineered G-alpha-q subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCC
 
 | | Cryo-EM structure of the CHA-bound mTAAR1 complex | | Descriptor: | CYCLOHEXYLAMMONIUM ION, Trace amine-associated receptor 1 | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC6
 
 | | Cryo-EM structure of the PEA-bound mTAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
4KQE
 
 | | The mutant structure of the human glycyl-tRNA synthetase E71G | | Descriptor: | GLYCEROL, Glycine--tRNA ligase | | Authors: | Qin, X, Hao, Z, Tian, Q, Zhang, Z, Zhou, C, Xie, W. | | Deposit date: | 2013-05-15 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.739 Å) | | Cite: | Large Conformational Changes of Insertion 3 in Human Glycyl-tRNA Synthetase (hGlyRS) during Catalysis
J.Biol.Chem., 291, 2016
|
|
7EN3
 
 | | Crystal structure of tubulin in complex with Tubulysin analogue TGL | | Descriptor: | (2S,4R)-5-(4-fluorophenyl)-2-methyl-4-[[2-[(1R,3R)-4-methyl-3-[5-methylhexyl-[(2S,3S)-3-methyl-2-[[(2R)-1-methylpiperidin-2-yl]carbonylamino]pentanoyl]amino]-1-oxidanyl-pentyl]-1,3-thiazol-4-yl]carbonylamino]pentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.643 Å) | | Cite: | The X-ray structure of tubulysin analogue TGL in complex with tubulin and three possible routes for the development of next-generation tubulysin analogues.
Biochem.Biophys.Res.Commun., 565, 2021
|
|
5UOP
 
 | | CRYSTAL STRUCTURE OF THE PROTOTYPE FOAMY VIRUS INTASOME WITH A 2- PYRIDINONE AMINAL INHIBITOR (COMPOUND 18) | | Descriptor: | (1S,2S,5R)-8'-[(3-chloro-4-fluorophenyl)methyl]-2'-[2-(2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl)ethyl]-6'-hydroxy-9',10'-dihydro-2'H-spiro[bicyclo[3.1.0]hexane-2,3'-imidazo[5,1-a][2,6]naphthyridine]-1',5',7'(8'H)-trione, GLYCEROL, INTEGRASE, ... | | Authors: | Klein, D.J. | | Deposit date: | 2017-02-01 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery and optimization of 2-pyridinone aminal integrase strand transfer inhibitors for the treatment of HIV.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5UOQ
 
 | | CRYSTAL STRUCTURE OF THE PROTOTYPE FOAMY VIRUS INTASOME WITH A 2- PYRIDINONE AMINAL INHIBITOR (COMPOUND 31) | | Descriptor: | (3R)-8-[(3-chloro-4-fluorophenyl)methyl]-6-hydroxy-1,5,7-trioxo-1,2',3',5,7,8,9,10-octahydro-2H-spiro[imidazo[5,1-a][2,6]naphthyridine-3,1'-indene]-7'-carbonitrile, GLYCEROL, INTEGRASE, ... | | Authors: | Klein, D.J. | | Deposit date: | 2017-02-01 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery and optimization of 2-pyridinone aminal integrase strand transfer inhibitors for the treatment of HIV.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6X4H
 
 | | Sortilin-Progranulin Interaction With Compound 24 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-methyl-N-(6-phenoxypyridine-3-carbonyl)-L-leucine, GLYCEROL, ... | | Authors: | Parthasarathy, G, Soisson, S.M, Klein, D. | | Deposit date: | 2020-05-22 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of potent inhibitors of the sortilin-progranulin interaction.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
4ZTJ
 
 | | Crystal Structure of the Prototype Foamy Virus Intasome with a 2-Pyridinone Aminal Inhibitor | | Descriptor: | (1R,2S,5R)-8'-(3-chloro-4-fluorobenzyl)-6'-hydroxy-1-(hydroxymethyl)-2'-methyl-9',10'-dihydro-2'H-spiro[bicyclo[3.1.0]hexane-2,3'-imidazo[5,1-a][2,6]naphthyridine]-1',5',7'(8'H)-trione, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | Authors: | Klein, D.J, Patel, S. | | Deposit date: | 2015-05-14 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Discovery of 2-Pyridinone Aminals: A Prodrug Strategy to Advance a Second Generation of HIV-1 Integrase Strand Transfer Inhibitors.
J.Med.Chem., 58, 2015
|
|
