5Y93
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[5-[(5-bromanyl-2-methoxy-phenyl)sulfonylamino]-3-methyl-1,2-benzoxazol-6-yl]oxy]-N-(2-morpholin-4-ylethyl)ethanamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5DDU
 
 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with PMP | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CrmG, GLYCEROL, ... | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
5DDS
 
 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with PLP | | Descriptor: | ACETIC ACID, CrmG, GLYCEROL, ... | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
5XXH
 
 | | Crystal Structure Analysis of the CBP | | Descriptor: | (3S)-1-[2-(3-ethanoylindol-1-yl)ethanoyl]piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, CREB-binding protein, ... | | Authors: | Xiang, Q, Zhang, Y, Wang, C, Song, M. | | Deposit date: | 2017-07-04 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and optimization of 1-(1H-indol-1-yl)ethanone derivatives as CBP/EP300 bromodomain inhibitors for the treatment of castration-resistant prostate cancer.
Eur J Med Chem, 147, 2018
|
|
7S8O
 
 | | CryoEM structure of Gi-coupled MRGPRX2 with small molecule agonist (R)-Zinc-3573 | | Descriptor: | (3R)-N,N-dimethyl-1-[(8S)-5-phenylpyrazolo[1,5-a]pyrimidin-7-yl]pyrrolidin-3-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cao, C, Fay, J.F, Gumpper, R.H, Roth, B.L. | | Deposit date: | 2021-09-18 | | Release date: | 2021-11-17 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Structure, function and pharmacology of human itch GPCRs.
Nature, 600, 2021
|
|
7S8L
 
 | | CryoEM structure of Gq-coupled MRGPRX2 with peptide agonist Cortistatin-14 | | Descriptor: | Cortistatin 14, Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cao, C, Fay, J.F, Gumpper, R.H, Roth, B.L. | | Deposit date: | 2021-09-18 | | Release date: | 2021-11-17 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structure, function and pharmacology of human itch GPCRs.
Nature, 600, 2021
|
|
7S8M
 
 | | CryoEM structure of Gi-coupled MRGPRX2 with peptide agonist Cortistatin-14 | | Descriptor: | Cortistatin 14, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cao, C, Fay, J.F, Gumpper, R.H, Roth, B.L. | | Deposit date: | 2021-09-18 | | Release date: | 2021-11-17 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structure, function and pharmacology of human itch GPCRs.
Nature, 600, 2021
|
|
7S8N
 
 | | CryoEM structure of Gq-coupled MRGPRX2 with small molecule agonist (R)-Zinc-3573 | | Descriptor: | (3R)-N,N-dimethyl-1-[(8S)-5-phenylpyrazolo[1,5-a]pyrimidin-7-yl]pyrrolidin-3-amine, Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cao, C, Fay, J.F, Gumpper, R.H, Roth, B.L. | | Deposit date: | 2021-09-18 | | Release date: | 2021-11-17 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure, function and pharmacology of human itch GPCRs.
Nature, 600, 2021
|
|
7S8P
 
 | | CryoEM structure of Gq-coupled MRGPRX4 with small molecule agonist MS47134 | | Descriptor: | Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cao, C, Fay, J.F, Gumpper, R.H, Roth, B.L. | | Deposit date: | 2021-09-18 | | Release date: | 2021-11-17 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure, function and pharmacology of human itch GPCRs.
Nature, 600, 2021
|
|
6ZY9
 
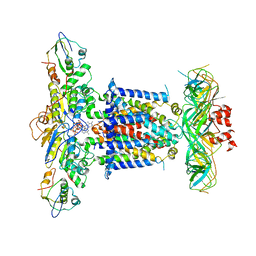 | | Cryo-EM structure of MlaFEDB in complex with AMP-PNP | | Descriptor: | ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, MAGNESIUM ION, ... | | Authors: | Dong, C.J, Dong, H.H. | | Deposit date: | 2020-07-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZY4
 
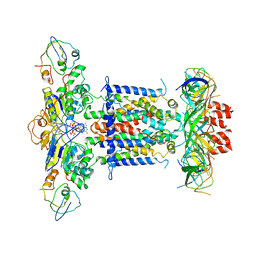 | | Cryo-EM structure of MlaFEDB in complex with ADP | | Descriptor: | ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Dong, C.J, Dong, H.H. | | Deposit date: | 2020-07-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZY3
 
 | | Cryo-EM structure of MlaFEDB in complex with phospholipid | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, ... | | Authors: | Dong, C.J, Dong, H.H. | | Deposit date: | 2020-07-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZY2
 
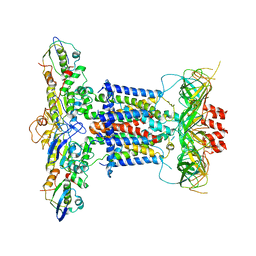 | | Cryo-EM structure of apo MlaFEDB | | Descriptor: | ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, Toluene tolerance protein Ttg2A, ... | | Authors: | Dong, C.J, Dong, H.H. | | Deposit date: | 2020-07-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5H19
 
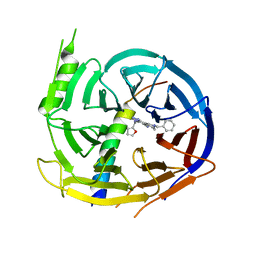 | | EED in complex with PRC2 allosteric inhibitor EED162 | | Descriptor: | 5-(furan-2-ylmethylamino)-9-(phenylmethyl)-8,10-dihydro-7H-[1,2,4]triazolo[3,4-a][2,7]naphthyridine-6-carbonitrile, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
5H24
 
 | | EED in complex with PRC2 allosteric inhibitor compound 8 | | Descriptor: | 5-(furan-2-ylmethylamino)-[1,2,4]triazolo[4,3-a]pyridine-6-carbonitrile, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-14 | | Release date: | 2017-01-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of First-in-Class, Potent, and Orally Bioavailable Embryonic Ectoderm Development (EED) Inhibitor with Robust Anticancer Efficacy
J. Med. Chem., 60, 2017
|
|
5GSA
 
 | | EED in complex with an allosteric PRC2 inhibitor | | Descriptor: | Histone-lysine N-methyltransferase EZH2, N-(furan-2-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of
Nat. Chem. Biol., 13, 2017
|
|
5GUT
 
 | | The crystal structure of mouse DNMT1 (731-1602) mutant - N1248A | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Chen, S.J, Ye, F. | | Deposit date: | 2016-08-31 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Biochemical Studies and Molecular Dynamic Simulations Reveal the Molecular Basis of Conformational Changes in DNA Methyltransferase-1.
ACS Chem. Biol., 13, 2018
|
|
5GUV
 
 | | The crystal structure of mouse DNMT1 (731-1602) mutant - R1279D | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, ZINC ION | | Authors: | Ye, F, Chen, S.J. | | Deposit date: | 2016-08-31 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.078 Å) | | Cite: | Biochemical Studies and Molecular Dynamic Simulations Reveal the Molecular Basis of Conformational Changes in DNA Methyltransferase-1.
ACS Chem. Biol., 13, 2018
|
|
4W5I
 
 | | Crystal structure of human tankyrase 2 in complex with 1-methyl-7-phenyl-1,2,3,4,5,6-hexahydro-1,6- naphthyridin-5-one | | Descriptor: | 1-methyl-7-phenyl-2,3,4,6-tetrahydro-1,6-naphthyridin-5(1H)-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design, synthesis and evaluation in vitro of arylnaphthyridinones, arylpyridopyrimidinones and their tetrahydro derivatives as inhibitors of the tankyrases.
Bioorg.Med.Chem., 23, 2015
|
|
9KQK
 
 | | Crystal structure of HSA in complex with AmpHecy | | Descriptor: | 2-[[6-[5-[(~{E})-2-[1-[2-[2-(2-methoxyethoxy)ethoxy]ethyl]pyridin-1-ium-4-yl]ethenyl]thiophen-2-yl]naphthalen-2-yl]-methyl-amino]ethanol, Albumin | | Authors: | Chen, X, Shi, M.W, Ge, Y.H, Fang, B, Li, L. | | Deposit date: | 2024-11-26 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Fluorogen-Activating Human Serum Albumin for Mitochondrial Nanoscale Imaging.
Adv Mater, 2025
|
|
6M3M
 
 | |
4FZO
 
 | |
4FZP
 
 | |
6S8N
 
 | | Cryo-EM structure of LptB2FGC in complex with lipopolysaccharide | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, Inner membrane protein yjgQ, ... | | Authors: | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
6S8H
 
 | | Cryo-EM structure of LptB2FG in complex with LPS | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
