7LY3
 
 | |
3OBR
 
 | |
3K5J
 
 | |
9IK2
 
 | | The co-crystal structure of SARS-CoV-2 Mpro in complex with compound H109 | | Descriptor: | 3C-like proteinase, tert-butyl N-[(2S)-1-[[(2S)-1-[[(2S)-1-azanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | Feng, Y, Zheng, W.Y, Han, P, Fu, L.F, Qi, J.X. | | Deposit date: | 2024-06-26 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided discovery of a small molecule inhibitor of SARS-CoV-2 main protease with potent in vitro and in vivo antiviral activities
To Be Published
|
|
4WT2
 
 | | Co-crystal Structure of MDM2 in Complex with AM-7209 | | Descriptor: | 4-({[(3R,5R,6S)-1-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(4-chloro-3-fluorophenyl)-5-(3-chlorophenyl)-3-methyl-2-oxopiperidin-3-yl]acetyl}amino)-2-methoxybenzoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-10-30 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery of AM-7209, a Potent and Selective 4-Amidobenzoic Acid Inhibitor of the MDM2-p53 Interaction.
J.Med.Chem., 57, 2014
|
|
3NMZ
 
 | | Crystal structure of APC complexed with Asef | | Descriptor: | APC variant protein, Rho guanine nucleotide exchange factor 4 | | Authors: | Zhang, Z, Chen, L, Gao, L, Lin, K, Wu, G. | | Deposit date: | 2010-06-23 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for the recognition of Asef by adenomatous polyposis coli.
Cell Res., 22, 2012
|
|
3NMX
 
 | | Crystal structure of APC complexed with Asef | | Descriptor: | APC variant protein, Rho guanine nucleotide exchange factor 4 | | Authors: | Zhang, Z, Chen, L, Gao, L, Lin, K, Wu, G. | | Deposit date: | 2010-06-22 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the recognition of Asef by adenomatous polyposis coli.
Cell Res., 22, 2012
|
|
5T0L
 
 | | Crystal structure of Toxoplasma gondii TS-DHFR complexed with NADPH, dUMP, PDDF and 5-(4-(3,4-dichlorophenyl)piperazin-1-yl)pyrimidine-2,4-diamine (TRC-15) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-[4-(3,4-dichlorophenyl)piperazin-1-yl]pyrimidine-2,4-diamine, ... | | Authors: | Thomas, S.B, Chen, Z, Lu, H, Li, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Discovery of Potent and Selective Leads against Toxoplasma gondii Dihydrofolate Reductase via Structure-Based Design.
ACS Med Chem Lett, 7, 2016
|
|
6LCU
 
 | |
7WZX
 
 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | Descriptor: | (2~{S},4~{S},6~{R})-2-[(2~{S},3~{R},5~{S},6~{R})-3,5-bis(methylamino)-2,4,6-tris(oxidanyl)cyclohexyl]oxy-6-methyl-4-oxidanyl-oxan-3-one, 4Fe-4S cluster-binding domain-containing protein, GLYCEROL, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2022-02-19 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.980013 Å) | | Cite: | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7X0B
 
 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | Descriptor: | 4Fe-4S cluster-binding domain-containing protein, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2022-02-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02027535 Å) | | Cite: | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7WZV
 
 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | Descriptor: | (1~{S},2~{R},4~{S},5~{R})-2,4-bis(methylamino)-6-[(2~{S},3~{R},4~{S},6~{R})-6-methyl-3,4-bis(oxidanyl)oxan-2-yl]oxy-cyclohexane-1,3,5-triol, 1,2-ETHANEDIOL, 4Fe-4S cluster-binding domain-containing protein, ... | | Authors: | Zhou, J.H, Hou, X.L. | | Deposit date: | 2022-02-19 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.899313 Å) | | Cite: | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
8IUF
 
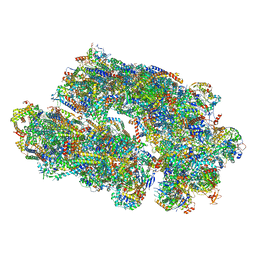 | | Cryo-EM structure of Euglena gracilis super-complex I+III2+IV, composite | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Wu, M.C, Tian, H.T, He, Z.X, Hu, Y.Q, Zhou, L. | | Deposit date: | 2023-03-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Euglena's atypical respiratory chain adapts to the discoidal cristae and flexible metabolism.
Nat Commun, 15, 2024
|
|
8J9I
 
 | | Cryo-EM structure of Euglena gracilis complex I, turnover state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Wu, M.C, He, Z.X, Tian, H.T, Hu, Y.Q, Han, F.Z, Zhou, L. | | Deposit date: | 2023-05-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Euglena's atypical respiratory chain adapts to the discoidal cristae and flexible metabolism.
Nat Commun, 15, 2024
|
|
8J9J
 
 | | Cryo-EM structure of Euglena gracilis complex I, NADH state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Wu, M.C, He, Z.X, Tian, H.T, Hu, Y.Q, Han, F.Z, Zhou, L. | | Deposit date: | 2023-05-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Euglena's atypical respiratory chain adapts to the discoidal cristae and flexible metabolism.
Nat Commun, 15, 2024
|
|
8IUJ
 
 | | Cryo-EM structure of Euglena gracilis super-complex III2+IV2, composite | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Wu, M.C, Tian, H.T, He, Z.X, Hu, Y.Q, Zhou, L. | | Deposit date: | 2023-03-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Euglena's atypical respiratory chain adapts to the discoidal cristae and flexible metabolism.
Nat Commun, 15, 2024
|
|
8J9H
 
 | | Cryo-EM structure of Euglena gracilis respiratory complex I, deactive state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Wu, M.C, He, Z.X, Tian, H.T, Hu, Y.Q, Han, F.Z, Zhou, L. | | Deposit date: | 2023-05-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Euglena's atypical respiratory chain adapts to the discoidal cristae and flexible metabolism.
Nat Commun, 15, 2024
|
|
3NMW
 
 | | Crystal structure of armadillo repeats domain of APC | | Descriptor: | APC variant protein, SULFATE ION | | Authors: | Zhang, Z, Chen, L, Gao, L, Lin, K, Wu, G. | | Deposit date: | 2010-06-22 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Asef by adenomatous polyposis coli.
Cell Res., 22, 2012
|
|
4ERE
 
 | | crystal structure of MDM2 (17-111) in complex with compound 23 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(3R,5R,6S)-1-[(2S)-1-tert-butoxy-1-oxobutan-2-yl]-5-(3-chlorophenyl)-6-(4-chlorophenyl)-2-oxopiperidin-3-yl]acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
4JVE
 
 | | Co-crystal structure of MDM2 with inhibitor (2R,3E)-2-[(2S,3R,6S)-2,3-bis(4-chlorophenyl)-6-(4-fluorobenzyl)-5-oxomorpholin-4-yl]pent-3-enoic acid | | Descriptor: | (2R,3E)-2-[(2S,3R,6S)-2,3-bis(4-chlorophenyl)-6-(4-fluorobenzyl)-5-oxomorpholin-4-yl]pent-3-enoic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4JV7
 
 | | Co-crystal structure of MDM2 with inhibitor (2S,5R,6S)-2-benzyl-5,6-bis(4-bromophenyl)-4-methylmorpholin-3-one | | Descriptor: | (2S,5R,6S)-2-benzyl-5,6-bis(4-bromophenyl)-4-methylmorpholin-3-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4JVR
 
 | | Co-crystal structure of MDM2 with inhibitor (2'S,3R,4'S,5'R)-N-(2-aminoethyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide | | Descriptor: | (2'S,3R,4'S,5'R)-N-(2-aminoethyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-26 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4JV9
 
 | | Co-crystal structure of MDM2 with inhibitor (2S,5R,6S)-2-benzyl-5,6-bis(4-chlorophenyl)-4-methylmorpholin-3-one | | Descriptor: | (2S,5R,6S)-2-benzyl-5,6-bis(4-chlorophenyl)-4-methylmorpholin-3-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4ERF
 
 | | crystal structure of MDM2 (17-111) in complex with compound 29 (AM-8553) | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, {(3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-1-[(2S,3S)-2-hydroxypentan-3-yl]-3-methyl-2-oxopiperidin-3-yl}acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
7MU5
 
 | | Human DCTPP1 bound to Triptolide | | Descriptor: | MAGNESIUM ION, dCTP pyrophosphatase 1, triptolide | | Authors: | Hauk, G, Berger, J.M. | | Deposit date: | 2021-05-14 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Triptolide sensitizes cancer cells to nucleoside DNA methyltransferase inhibitors through inhibition of DCTPP1 mediated cell-intrinsic resistance
To Be Published
|
|
